Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs - PubMed (original) (raw)
B Weil, R Wellenreuther, J Gassenhuber, S Glassl, W Ansorge, M Böcher, H Blöcker, S Bauersachs, H Blum, J Lauber, A Düsterhöft, A Beyer, K Köhrer, N Strack, H W Mewes, B Ottenwälder, B Obermaier, J Tampe, D Heubner, R Wambutt, B Korn, M Klein, A Poustka
Affiliations
- PMID: 11230166
- PMCID: PMC311072
- DOI: 10.1101/gr.gr1547r
Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs
S Wiemann et al. Genome Res. 2001 Mar.
Abstract
With the complete human genomic sequence being unraveled, the focus will shift to gene identification and to the functional analysis of gene products. The generation of a set of cDNAs, both sequences and physical clones, which contains the complete and noninterrupted protein coding regions of all human genes will provide the indispensable tools for the systematic and comprehensive analysis of protein function to eventually understand the molecular basis of man. Here we report the sequencing and analysis of 500 novel human cDNAs containing the complete protein coding frame. Assignment to functional categories was possible for 52% (259) of the encoded proteins, the remaining fraction having no similarities with known proteins. By aligning the cDNA sequences with the sequences of the finished chromosomes 21 and 22 we identified a number of genes that either had been completely missed in the analysis of the genomic sequences or had been wrongly predicted. Three of these genes appear to be present in several copies. We conclude that full-length cDNA sequencing continues to be crucial also for the accurate identification of genes. The set of 500 novel cDNAs, and another 1000 full-coding cDNAs of known transcripts we have identified, adds up to cDNA representations covering 2%--5 % of all human genes. We thus substantially contribute to the generation of a gene catalog, consisting of both full-coding cDNA sequences and clones, which should be made freely available and will become an invaluable tool for detailed functional studies.
Figures
Figure 1
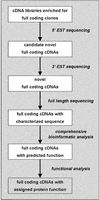
Flow of clones, sequences, and information in the German cDNA Consortium. 5′ EST sequences were systematically generated from the clones of 384-well microtiter plates and analyzed for hits in public databases. Clones with novel sequences were 3′-EST sequenced and these ESTs were analyzed again for novelty. Clones of uncharacterized transcripts were reported back to the sequencers who then did the full-length sequencing of cDNAs. The final sequence was analyzed comprehensively with bioinformatic tools and the outputs were evaluated manually. The clones feed functional analysis projects that take advantage of the clone resources generated.
Figure 2
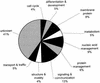
Functional classification of proteins encoded by the cDNAs. The deduced proteins were grouped into 10 functional categories based on sequence similarity with proteins of known function. The fraction of the 500 cDNAs grouped into the respective categories is indicated.
Figure 3
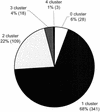
Representation of cDNAs in the UniGene data set (Build 105). Every cDNA was aligned with the UniGene data set to identify the number of EST clusters that was hit/joined with a given cDNA. The fraction and the total number (in parentheses) of the cDNAs are given for the varying numbers of clusters being hit.
Figure 4
Three UniGene clusters are joined when aligned with the cDNA sequence DKFZp434B0435. The bar on top of the scale represents the cDNA with the open reading frame drawn as an open box. The bars below the scale represent the position and size (in bp) of the three UniGene clusters that are joined by the cDNA sequence. The accession nos. of representative sequences of the respective UniGene clusters are given below the bars.
Figure 5
Multiple sequence alignment of cDNA DKFZp434P211 with POM121-related 1 (accession no. D87002) and sequences from chromosome 22 demonstrate the presence of a cluster of POM121-related genes. The individual genomic sequences were named after the start of the first exon relative to the cDNA: The open reading frame (ORF) was defined according to the predicted protein of the cDNA and of POM121-related 1. Genes located on the plus and minus strands of chromosome 22 are indicated with + and −, respectively. The cDNA sequence of DKFZp434P211 was taken as reference; identical residues in other sequences are indicated with a dot, residues deviating from the consensus are printed. Asterisks (*) indicate stop codons. The genomic sequences 2850458 and 2871777 are in italics because these copies deviate from the other copies by a premature stop or frame shifts and a large insertion, respectively, and are probably not expressed. In these two gene copies the initiator ATG is mutated. Dashes (-) were inserted by the software (
CLUSTAL
) to optimize the alignment.
Similar articles
- Characterization of cDNA clones selected by the GeneMark analysis from size-fractionated cDNA libraries from human brain.
Hirosawa M, Nagase T, Ishikawa K, Kikuno R, Nomura N, Ohara O. Hirosawa M, et al. DNA Res. 1999 Oct 29;6(5):329-36. doi: 10.1093/dnares/6.5.329. DNA Res. 1999. PMID: 10574461 - Comparative genomic analysis of 1047 completely sequenced cDNAs from an Arabidopsis-related model halophyte, Thellungiella halophila.
Taji T, Komatsu K, Katori T, Kawasaki Y, Sakata Y, Tanaka S, Kobayashi M, Toyoda A, Seki M, Shinozaki K. Taji T, et al. BMC Plant Biol. 2010 Nov 24;10:261. doi: 10.1186/1471-2229-10-261. BMC Plant Biol. 2010. PMID: 21106055 Free PMC article. - Characterization of 954 bovine full-CDS cDNA sequences.
Harhay GP, Sonstegard TS, Keele JW, Heaton MP, Clawson ML, Snelling WM, Wiedmann RT, Van Tassell CP, Smith TP. Harhay GP, et al. BMC Genomics. 2005 Nov 23;6:166. doi: 10.1186/1471-2164-6-166. BMC Genomics. 2005. PMID: 16305752 Free PMC article. - Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones.
Nakajima D, Okazaki N, Yamakawa H, Kikuno R, Ohara O, Nagase T. Nakajima D, et al. DNA Res. 2002 Jun 30;9(3):99-106. doi: 10.1093/dnares/9.3.99. DNA Res. 2002. PMID: 12168954 Review. - Transcriptome analyses of human genes and applications for proteome analyses.
Suzuki Y, Sugano S. Suzuki Y, et al. Curr Protein Pept Sci. 2006 Apr;7(2):147-63. doi: 10.2174/138920306776359795. Curr Protein Pept Sci. 2006. PMID: 16611140 Review.
Cited by
- Large-scale sequencing based on full-length-enriched cDNA libraries in pigs: contribution to annotation of the pig genome draft sequence.
Uenishi H, Morozumi T, Toki D, Eguchi-Ogawa T, Rund LA, Schook LB. Uenishi H, et al. BMC Genomics. 2012 Nov 15;13:581. doi: 10.1186/1471-2164-13-581. BMC Genomics. 2012. PMID: 23150988 Free PMC article. - Automated production of recombinant human proteins as resource for proteome research.
Kohl T, Schmidt C, Wiemann S, Poustka A, Korf U. Kohl T, et al. Proteome Sci. 2008 Jan 28;6:4. doi: 10.1186/1477-5956-6-4. Proteome Sci. 2008. PMID: 18226205 Free PMC article. - Identification of cellular proteins that interact with human cytomegalovirus immediate-early protein 1 by protein array assay.
Martínez FP, Tang Q. Martínez FP, et al. Viruses. 2013 Dec 31;6(1):89-105. doi: 10.3390/v6010089. Viruses. 2013. PMID: 24385082 Free PMC article. - Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold.
Manjasetty BA, Büssow K, Fieber-Erdmann M, Roske Y, Gobom J, Scheich C, Götz F, Niesen FH, Heinemann U. Manjasetty BA, et al. Protein Sci. 2006 Apr;15(4):914-20. doi: 10.1110/ps.052037006. Epub 2006 Mar 7. Protein Sci. 2006. PMID: 16522806 Free PMC article. - From ORFeome to biology: a functional genomics pipeline.
Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sültmann H, Poustka A. Wiemann S, et al. Genome Res. 2004 Oct;14(10B):2136-44. doi: 10.1101/gr.2576704. Genome Res. 2004. PMID: 15489336 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. - PubMed
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. - PubMed
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases