A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation - PubMed (original) (raw)
Comparative Study
A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation
M R Nelson et al. Genome Res. 2001 Mar.
Abstract
Recent advances in genome research have accelerated the process of locating candidate genes and the variable sites within them and have simplified the task of genotype measurement. The development of statistical and computational strategies to utilize information on hundreds -- soon thousands -- of variable loci to investigate the relationships between genome variation and phenotypic variation has not kept pace, particularly for quantitative traits that do not follow simple Mendelian patterns of inheritance. We present here the combinatorial partitioning method (CPM) that examines multiple genes, each containing multiple variable loci, to identify partitions of multilocus genotypes that predict interindividual variation in quantitative trait levels. We illustrate this method with an application to plasma triglyceride levels collected on 188 males, ages 20--60 yr, ascertained without regard to health status, from Rochester, Minnesota. Genotype information included measurements at 18 diallelic loci in six coronary heart disease--candidate susceptibility gene regions: APOA1--C3--A4, APOB, APOE, LDLR, LPL, and PON1. To illustrate the CPM, we evaluated all possible partitions of two-locus genotypes into two to nine partitions (approximately 10(6) evaluations). We found that many combinations of loci are involved in sets of genotypic partitions that predict triglyceride variability and that the most predictive sets show nonadditivity. These results suggest that traditional methods of building multilocus models that rely on statistically significant marginal, single-locus effects, may fail to identify combinations of loci that best predict trait variability. The CPM offers a strategy for exploring the high-dimensional genotype state space so as to predict the quantitative trait variation in the population at large that does not require the conditioning of the analysis on a prespecified genetic model.
Figures
Figure 1
The three steps that constitute the combinatorial partitioning method.
Figure 2
A depiction of the combinatorial partitioning method applied two variable loci at a time (m = 2) over a range of k.
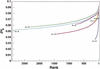
Figure 3
Plot of the proportion of variability explained by the 7710 retained sets of genotypic partitions. The sets are sorted by the proportion of variability explained and connected by a colored line corresponding to the number of partitions in each set.
Figure 4
Plot of the proportion of variability explained by the same sets shown in Figure 3, after grouping by the pairs of variable loci included in each set and sorting groups by the proportion of variability explained by the best partition for each group.
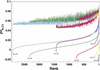
Figure 5
Plot of the cross-validated proportion of variability explained by the 7710 retained sets of genotypic partitions. The sets are sorted by the proportion of variability explained and connected by a colored line corresponding to the number of partitions in each set (smooth, nondecreasing lines). The proportion of variability explained for each set before cross-validation is shown by the jagged lines of corresponding colors.
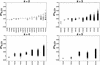
Figure 6
Plot of the proportion of cross-validated variability explained by the same sets shown in Figure 5, after grouping by the pairs of variable loci included in each set and sorting groups by the proportion of variability explained by the best partition for each group.
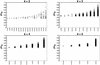
Figure 7
The three selected sets of genotypic partitions with the greatest proportion of cross-validated variability explained are represented by a 3 × 3 grid of nine two-locus genotypes with shading to represent the partition each genotype belongs to. Below each partition is a smoothed histogram showing the ln Trig distribution within each partition (indicated by shading) and the mean and sample size of each partition.
Similar articles
- Contrasting multi-site genotypic distributions among discordant quantitative phenotypes: the APOA1/C3/A4/A5 gene cluster and cardiovascular disease risk factors.
Payseur BA, Clark AG, Hixson J, Boerwinkle E, Sing CF. Payseur BA, et al. Genet Epidemiol. 2006 Sep;30(6):508-18. doi: 10.1002/gepi.20163. Genet Epidemiol. 2006. PMID: 16800005 - A genome-wide association study identified loci for yield component traits in sugarcane (Saccharum spp.).
Barreto FZ, Rosa JRBF, Balsalobre TWA, Pastina MM, Silva RR, Hoffmann HP, de Souza AP, Garcia AAF, Carneiro MS. Barreto FZ, et al. PLoS One. 2019 Jul 18;14(7):e0219843. doi: 10.1371/journal.pone.0219843. eCollection 2019. PLoS One. 2019. PMID: 31318931 Free PMC article. - Linkage analysis of quantitative traits in randomly ascertained pedigrees: comparison of penetrance-based and variance component analysis.
Göring HH, Williams JT, Blangero J. Göring HH, et al. Genet Epidemiol. 2001;21 Suppl 1:S783-8. doi: 10.1002/gepi.2001.21.s1.s783. Genet Epidemiol. 2001. PMID: 11793778 - Understanding and predicting complex traits: knowledge from cattle.
Kemper KE, Goddard ME. Kemper KE, et al. Hum Mol Genet. 2012 Oct 15;21(R1):R45-51. doi: 10.1093/hmg/dds332. Epub 2012 Aug 16. Hum Mol Genet. 2012. PMID: 22899652 Review.
Cited by
- A novel bayesian graphical model for genome-wide multi-SNP association mapping.
Zhang Y. Zhang Y. Genet Epidemiol. 2012 Jan;36(1):36-47. doi: 10.1002/gepi.20661. Epub 2011 Nov 29. Genet Epidemiol. 2012. PMID: 22127647 Free PMC article. - Associations among race/ethnicity, ApoC-III genotypes, and lipids in HIV-1-infected individuals on antiretroviral therapy.
Foulkes AS, Wohl DA, Frank I, Puleo E, Restine S, Wolfe ML, Dube MP, Tebas P, Reilly MP. Foulkes AS, et al. PLoS Med. 2006 Mar;3(3):e52. doi: 10.1371/journal.pmed.0030052. PLoS Med. 2006. PMID: 16417409 Free PMC article. - A combinatorial approach for detecting gene-gene interaction using multiple traits of Genetic Analysis Workshop 16 rheumatoid arthritis data.
Cui X, Sha Q, Zhang S, Chen HS. Cui X, et al. BMC Proc. 2009 Dec 15;3 Suppl 7(Suppl 7):S43. doi: 10.1186/1753-6561-3-s7-s43. BMC Proc. 2009. PMID: 20018035 Free PMC article. - Candidate genes and their interactions with other genetic/environmental risk factors in the etiology of schizophrenia.
Prasad KM, Talkowski ME, Chowdari KV, McClain L, Yolken RH, Nimgaonkar VL. Prasad KM, et al. Brain Res Bull. 2010 Sep 30;83(3-4):86-92. doi: 10.1016/j.brainresbull.2009.08.023. Epub 2009 Sep 1. Brain Res Bull. 2010. PMID: 19729054 Free PMC article. Review. - The challenge for genetic epidemiologists: how to analyze large numbers of SNPs in relation to complex diseases.
Heidema AG, Boer JM, Nagelkerke N, Mariman EC, van der A DL, Feskens EJ. Heidema AG, et al. BMC Genet. 2006 Apr 21;7:23. doi: 10.1186/1471-2156-7-23. BMC Genet. 2006. PMID: 16630340 Free PMC article.
References
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Monterey, CA: Wadsworth & Brooks/Cole; 1984.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous