Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays - PubMed (original) (raw)
Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays
M Raitio et al. Genome Res. 2001 Mar.
Abstract
An increasing number of single nucleotide polymorphisms (SNPs) on the Y chromosome are being identified. To utilize the full potential of the SNP markers in population genetic studies, new genotyping methods with high throughput are required. We describe a microarray system based on the minisequencing single nucleotide primer extension principle for multiplex genotyping of Y-chromosomal SNP markers. The system was applied for screening a panel of 25 Y-chromosomal SNPs in a unique collection of samples representing five Finno--Ugric populations. The specific minisequencing reaction provides 5-fold to infinite discrimination between the Y-chromosomal genotypes, and the microarray format of the system allows parallel and simultaneous analysis of large numbers of SNPs and samples. In addition to the SNP markers, five Y-chromosomal microsatellite loci were typed. Altogether 10,000 genotypes were generated to assess the genetic diversity in these population samples. Six of the 25 SNP markers (M9, Tat, SRY10831, M17, M12, 92R7) were polymorphic in the analyzed populations, yielding six distinct SNP haplotypes. The microsatellite data were used to study the genetic structure of two major SNP haplotypes in the Finns and the Saami in more detail. We found that the most common haplotypes are shared between the Finns and the Saami, and that the SNP haplotypes show regional differences within the Finns and the Saami, which supports the hypothesis of two separate settlement waves to Finland.
Figures
Figure 1
The geographic origin of the sampled Finno–Ugric-speaking populations and the frequencies of Y-chromosomal haplotypes formed by SNPs in these populations. For haplotype designation and nomenclature as well as the population frequencies of the observed haplotypes, see Table 5.
Figure 2
Result from validation of the microarray-based genotyping system. Two mixtures of 25 synthetic oligonucleotide templates corresponding to one of the alleles at each SNP site were analyzed in minisequencing reactions on the microarrays to verify that both alleles of each SNP marker would be detected. (Left panel) The fluorescence image obtained from four detection reactions (A, C, G, T) with TAMRA-labeled ddNTPs. (Right panel) Order of the SNP markers on the array with the nucleotide variation at each site. The asterisk indicates those alleles that are detected from complementary DNA strands compared to earlier published literature.
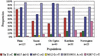
Figure 3
Allele frequencies of six polymorphic Y-chromosomal SNP markers.
Similar articles
- [A minisequencing technique for genotyping 12 Y-SNP and its genetic polymorphisms].
Du H, Zhang L, Zhou B, Zhang HJ, Liang WB, Shen YH. Du H, et al. Fa Yi Xue Za Zhi. 2006 Apr;22(2):125-9. Fa Yi Xue Za Zhi. 2006. PMID: 16850599 Chinese. - Genotyping single-nucleotide polymorphisms by minisequencing using tag arrays.
Lovmar L, Syvänen AC. Lovmar L, et al. Methods Mol Med. 2005;114:79-92. doi: 10.1385/1-59259-923-0:79. Methods Mol Med. 2005. PMID: 16156098 - Detecting imbalanced expression of SNP alleles by minisequencing on microarrays.
Liljedahl U, Fredriksson M, Dahlgren A, Syvänen AC. Liljedahl U, et al. BMC Biotechnol. 2004 Oct 22;4:24. doi: 10.1186/1472-6750-4-24. BMC Biotechnol. 2004. PMID: 15500681 Free PMC article. - Tag SNP selection for association studies.
Stram DO. Stram DO. Genet Epidemiol. 2004 Dec;27(4):365-74. doi: 10.1002/gepi.20028. Genet Epidemiol. 2004. PMID: 15372618 Review. - Array-based high-throughput DNA markers for crop improvement.
Gupta PK, Rustgi S, Mir RR. Gupta PK, et al. Heredity (Edinb). 2008 Jul;101(1):5-18. doi: 10.1038/hdy.2008.35. Epub 2008 May 7. Heredity (Edinb). 2008. PMID: 18461083 Review.
Cited by
- Microarray analysis of evolution of RNA viruses: evidence of circulation of virulent highly divergent vaccine-derived polioviruses.
Cherkasova E, Laassri M, Chizhikov V, Korotkova E, Dragunsky E, Agol VI, Chumakov K. Cherkasova E, et al. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9398-403. doi: 10.1073/pnas.1633511100. Epub 2003 Jul 23. Proc Natl Acad Sci U S A. 2003. PMID: 12878723 Free PMC article. - The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations.
Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Gölge M, Usanga E, Papiha SS, Cinnioğlu C, King R, Cavalli-Sforza L, Underhill PA, Villems R. Kivisild T, et al. Am J Hum Genet. 2003 Feb;72(2):313-32. doi: 10.1086/346068. Epub 2003 Jan 20. Am J Hum Genet. 2003. PMID: 12536373 Free PMC article. - DNA microarrays with PAMAM dendritic linker systems.
Benters R, Niemeyer CM, Drutschmann D, Blohm D, Wöhrle D. Benters R, et al. Nucleic Acids Res. 2002 Jan 15;30(2):E10. doi: 10.1093/nar/30.2.e10. Nucleic Acids Res. 2002. PMID: 11788736 Free PMC article. - Excavating Y-chromosome haplotype strata in Anatolia.
Cinnioğlu C, King R, Kivisild T, Kalfoğlu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA. Cinnioğlu C, et al. Hum Genet. 2004 Jan;114(2):127-48. doi: 10.1007/s00439-003-1031-4. Epub 2003 Oct 29. Hum Genet. 2004. PMID: 14586639 - Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization.
Chizhikov V, Wagner M, Ivshina A, Hoshino Y, Kapikian AZ, Chumakov K. Chizhikov V, et al. J Clin Microbiol. 2002 Jul;40(7):2398-407. doi: 10.1128/JCM.40.7.2398-2407.2002. J Clin Microbiol. 2002. PMID: 12089254 Free PMC article.
References
- Bianchi NO, Bailliet G, Bravi CM, Carnese RF, Rothhammer F, Martinez-Marignac VL, Pena SD. Origin of Amerindian Y-chromosomes as inferred by the analysis of six polymorphic markers. Am J Phys Anthropol. 1997;102:79–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources