Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements - PubMed (original) (raw)
doi: 10.1086/319506. Epub 2001 Feb 26.
K W Broman, N Matsumoto, V Calvari, G Gimelli, T Neumann, H Ohashi, L Voullaire, D Larizza, R Giorda, J L Weber, D H Ledbetter, O Zuffardi
Affiliations
- PMID: 11231899
- PMCID: PMC1275641
- DOI: 10.1086/319506
Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements
S Giglio et al. Am J Hum Genet. 2001 Apr.
Abstract
The olfactory receptor (OR)-gene superfamily is the largest in the mammalian genome. Several of the human OR genes appear in clusters with > or = 10 members located on almost all human chromosomes, and some chromosomes contain more than one cluster. We demonstrate, by experimental and in silico data, that unequal crossovers between two OR gene clusters in 8p are responsible for the formation of three recurrent chromosome macrorearrangements and a submicroscopic inversion polymorphism. The first two macrorearrangements are the inverted duplication of 8p, inv dup(8p), which is associated with a distinct phenotype, and a supernumerary marker chromosome, +der(8)(8p23.1pter), which is also a recurrent rearrangement and is associated with minor anomalies. We demonstrate that it is the reciprocal of the inv dup(8p). The third macrorearrangment is a recurrent 8p23 interstitial deletion associated with heart defect. Since inv dup(8p)s originate consistently in maternal meiosis, we investigated the maternal chromosomes 8 in eight mothers of subjects with inv dup(8p) and in the mother of one subject with +der(8), by means of probes included between the two 8p-OR gene clusters. All the mothers were heterozygous for an 8p submicroscopic inversion that was delimited by the 8p-OR gene clusters and was present, in heterozygous state, in 26% of a population of European descent. Thus, inversion heterozygosity may cause susceptibility to unequal recombination, leading to the formation of the inv dup(8p) or to its reciprocal product, the +der(8p). After the Yp inversion polymorphism, which is the preferential background for the PRKX/PRKY translocation in XX males and XY females, the OR-8p inversion is the second genomic polymorphism that confers susceptibility to the formation of common chromosome rearrangements. Accordingly, it may be possible to develop a profile of the individual risk of having progeny with chromosome rearrangements.
Figures
Figure 1
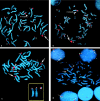
Inv dup(8p) breakpoint contigs. Middle, 8p ideogram. Shown in red is the region deleted in inv dup(8p); in blue, the region in single copy; and in brown, the region that can be totally or partially duplicated (Floridia et al. 1996). REPD = distal repeat; REPP = proximal repeat. All clones inside REPs give an OR FISH pattern. Top and bottom, STSs (in green) and BAC ends (SP6 and T7) anchored to the respective clones. Red/blue and blue/brown clones indicate the inv dup(8p) and the der(8p) breakpoints (see text). In the interstitial del(8p), red clones lie in the nondeleted region, red/blue clones indicate the distal breakpoint, and blue clones in REPD and between REPD and REPP up to 257o3 are deleted; all the proximal blue and brown clones are present. Colored squares indicate genes and pseudogenes found in the region. GATA4 = GATA-binding protein 4 gene; MYO (large square) = myosin gene; MYO (small squares) = myosin pseudogenes; OR = olfactory receptor genes, pseudogenes, and gene fragments; ANG2 = angiotensin II genes; and DEF = defensin genes (other defensins are located 100–150 kb proximal to REPD).
Figure 2
a and b, FISH with GS214h7 (a) and GS42i21 (b) in a prometaphase and a metaphase, respectively, from an inv dup(8p) subject. The two clones show a double set of signals (a) and a large signal (b) on the normal chromosome 8 (arrows) and a single smaller signal on the inv dup(8p)s (arrowheads). These BACs, which partially overlap and lie at the distal edge of REPD, also hybridize to other chromosomes (identified with numbers) containing OR gene clusters. c, FISH with GS42i21 in a metaphase from a subject with +der(8p). The signal pattern is the same as in a and b; the +der(8p) shows very large signals. Box, cut-out of chromosomes 8 and +der(8p), showing duplication of the red region of the contig in figure 1 (GS143g5) and monosomy of the contig’s blue region (GS77p24). d, FISH in a metaphase from the del(8p) subject with GS42i21 (green) and GS143g5 (red), the latter mapping distal to the two REPs. In the del(8p) (arrowhead), the green signal of the OR gene cluster is single and small, whereas, in the normal 8 (arrow), it is double and big. This FISH pattern is visible both in metaphase and interphase nuclei.
Figure 3
Metaphase from the mother of a subject with inv dup(8p), showing the normal (arrow) and the inverted (arrowhead) chromosomes 8 (magnified in the box). FISH was done with GS173o4 (red, D8S351), GS257o3 (green, D8S1130), both inside the inverted region (see fig. 1), and RP11-563o19 (yellow, D8S1733), ∼24 cM proximal to the inverted region.
Figure 4
Ideogram showing the mechanism of origin of the inv dup(8p) and of the +der(8p) at pachytene. Only the short arm of chromosome 8 is shown. a, At the first meiotic division (MI), the two maternal homologous chromosomes 8 (blue and black) undergo canonic recombination along the synapsed portion (in all informative inv dup(8p)s, part of the duplication region contains both maternal alleles [Floridia et al. 1996]). The region delimited by the two REPs (boxes) is inverted in the blue chromosome. Presence of heterozygous inversion leads to homologous synapsis interruption and to the refolding of the black chromosome, allowing intrachromatid synapsis and ectopic recombination between the two REPs (recombination is indicated by arrowheads). Red and green arrows show the orientation of the sequences inside each REP. As can be seen in fig. 1, two angiotensin II genes with the same orientation tel→cen, are present at the proximal and the distal portion of REPD. The orientation of REPP is hypothetical. Since a single FISH signal is visible in the inv dup(8p) with probes related to the OR gene clusters (see text), ectopic recombination should take place between the proximal portions of REPP and the distal one of REPD, thus leading to a very small OR cluster in the recombinant chromosome (hatched line). At MII, the recombinant chromosome (middle) shows a normal size gray OR cluster and a very small white/gray cluster. At anaphase II (right), when the centromere divides, the two chromatids, linked together, can join the opposite poles, provided that a breakage between the two centromeres occurs. If the breakage occurs at the level of one of the yellow lines, an inv dup(8p) is formed whose duplication size is determined by the position of this breakage. b, The same mechanism that produces the inv dup(8p) is responsible for the supernumerary der(8p) formation. The type of recombination is shown by arrowheads. The other chromosome 8 (blue) presumably undergo canonic recombination in the portion proximal to the inversion. This chromosome and the der(8p) segregate together in the oocyte. After anaphase II, the egg will contain a normal chromatid 8 and the der(8p). Formation of a neocentromere in the der(8p) will assure its preservation.
Similar articles
- Molecular characterization of inv dup del(8p): analysis of five cases.
Shimokawa O, Kurosawa K, Ida T, Harada N, Kondoh T, Miyake N, Yoshiura K, Kishino T, Ohta T, Niikawa N, Matsumoto N. Shimokawa O, et al. Am J Med Genet A. 2004 Jul 15;128A(2):133-7. doi: 10.1002/ajmg.a.30063. Am J Med Genet A. 2004. PMID: 15214003 - Molecular cytogenetic characterization of a unique and complex de novo 8p rearrangement.
Cooke SL, Northup JK, Champaige NL, Zinser W, Edwards PA, Lockhart LH, Velagaleti GV. Cooke SL, et al. Am J Med Genet A. 2008 May 1;146A(9):1166-72. doi: 10.1002/ajmg.a.32248. Am J Med Genet A. 2008. PMID: 18302246 - Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation.
Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, Zollino M, Neri G, Barber J, Wieczorek D, Rocchi M, Zuffardi O. Giglio S, et al. Am J Hum Genet. 2002 Aug;71(2):276-85. doi: 10.1086/341610. Epub 2002 Jun 10. Am J Hum Genet. 2002. PMID: 12058347 Free PMC article. - Clinical and cytogenetic findings in seven cases of inverted duplication of 8p with evidence of a telomeric deletion using fluorescence in situ hybridization.
Guo WJ, Callif-Daley F, Zapata MC, Miller ME. Guo WJ, et al. Am J Med Genet. 1995 Sep 11;58(3):230-6. doi: 10.1002/ajmg.1320580307. Am J Med Genet. 1995. PMID: 8533823 Review. - Molecular characterization of a new patient with a non-recurrent inv dup del 2q and review of the mechanisms for this rearrangement.
Vera-Carbonell A, López-Expósito I, Bafalliu JA, Ballesta-Martínez M, Glóver G, Llópis C, Moya-Quiles R, Suela J, Fernández A, Guillén-Navarro E. Vera-Carbonell A, et al. Am J Med Genet A. 2010 Oct;152A(10):2670-80. doi: 10.1002/ajmg.a.33613. Am J Med Genet A. 2010. PMID: 20799321 Review.
Cited by
- Structural Variation Evolution at the 15q11-q13 Disease-Associated Locus.
Paparella A, L'Abbate A, Palmisano D, Chirico G, Porubsky D, Catacchio CR, Ventura M, Eichler EE, Maggiolini FAM, Antonacci F. Paparella A, et al. Int J Mol Sci. 2023 Oct 31;24(21):15818. doi: 10.3390/ijms242115818. Int J Mol Sci. 2023. PMID: 37958807 Free PMC article. - Segmental duplications flank the multiple sclerosis locus on chromosome 17q.
Chen DC, Saarela J, Clark RA, Miettinen T, Chi A, Eichler EE, Peltonen L, Palotie A. Chen DC, et al. Genome Res. 2004 Aug;14(8):1483-92. doi: 10.1101/gr.2340804. Epub 2004 Jul 15. Genome Res. 2004. PMID: 15256512 Free PMC article. - Clinical Manifestations of Various Molecular Cytogenetic Variants of Eight Cases of "8p Inverted Duplication/Deletion Syndrome".
Yurchenko DA, Minzhenkova ME, Dadali EL, Markova ZG, Rudenskaya GE, Matyushchenko GN, Kanivets IV, Shilova NV. Yurchenko DA, et al. Biomedicines. 2022 Feb 28;10(3):567. doi: 10.3390/biomedicines10030567. Biomedicines. 2022. PMID: 35327368 Free PMC article. - Olfactory Receptor-Related Duplicons Mediate a Microdeletion at 11q13.2q13.4 Associated with a Syndromic Phenotype.
Wischmeijer A, Magini P, Giorda R, Gnoli M, Ciccone R, Cecconi L, Franzoni E, Mazzanti L, Romeo G, Zuffardi O, Seri M. Wischmeijer A, et al. Mol Syndromol. 2011 Jan;1(4):176-184. doi: 10.1159/000322054. Epub 2010 Nov 25. Mol Syndromol. 2011. PMID: 21373257 Free PMC article. - A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia.
Hardwick RJ, Machado LR, Zuccherato LW, Antolinos S, Xue Y, Shawa N, Gilman RH, Cabrera L, Berg DE, Tyler-Smith C, Kelly P, Tarazona-Santos E, Hollox EJ. Hardwick RJ, et al. Hum Mutat. 2011 Jul;32(7):743-50. doi: 10.1002/humu.21491. Epub 2011 May 5. Hum Mutat. 2011. PMID: 21387465 Free PMC article.
References
Electronic-Database Information
- Center for Genome Research, STS-Based Map of the Human Genome, http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Genome Database, http://gdbwww.gdb.org/ (for locus information and primer sequences)
- NIX-Identity Unk nown Nucleic Sequence, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/
References
- Brand-Arpon V, Rouquier S, Massa H, de Jong PJ, Ferraz C, Ioannou PA, Demaille JG, Trask BJ, Giorgi D (1999) A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13-q21 and 3p13. Genomics 56:98–110 - PubMed
- de Die-Smulders CE, Engelen JJ, Schrander-Stumpel CT, Govaerts LC, de Vries B, Vles JS, Wagemans A, Schijns-Fleuren S, Gillessen-Kaesbach G, Fryns JP (1995) Inversion duplication of the short arm of chromosome 8: clinical data on seven patients and review of the literature. Am J Med Genet 59:369–374 - PubMed
- Feldman GL, Weiss L, Phelan MC, Schroer RJ, Van Dyke DL (1993) Inverted duplication of 8p: ten new patients and review of the literature. Am J Med Genet 47:482–486 - PubMed
- Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, Croci G, Franchi F, Gilgenkrantz S, Grammatico P, Dalpra L, Wood S, Danesino C, Zuffardi O (1996) The same molecular mechanism at the maternal meiosis I produces mono and dicentric 8p duplications. Am J Hum Genet 58:785–796 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources