Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies - PubMed (original) (raw)
Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies
Q Mao et al. J Virol. 2001 Apr.
Abstract
When chronic hepatitis C virus (HCV) infections are complicated by acquisition of human immunodeficiency virus (HIV), liver disease appears to accelerate and serum levels of HCV RNA may rise. We hypothesized that HIV might affect the HCV quasispecies by decreasing both complexity (if HIV-induced immunosuppression lessens pressure for selecting HCV substitutions) and the ratio of nonsynonymous (d(N)) to synonymous (d(S)) substitutions, because d(N) may be lower (if there is less selective pressure). To test this hypothesis, we studied the evolution of HCV sequences in 10 persons with chronic HCV infection who seroconverted to HIV and, over the next 3 years, had slow or rapid progression of HIV-associated disease. From each subject, four serum specimens were selected with reference to HIV seroconversion: (i) more than 2 years prior, (ii) less than 2 years prior, (iii) less than 2 years after, and (iv) more than 2 years after. The HCV quasispecies in these specimens was characterized by generating clones containing 1 kb of cDNA that spanned the E1 gene and the E2 hypervariable region 1 (HVR1), followed by analysis of clonal frequencies (via electrophoretic migration) and nucleotide sequences. We examined 1,320 cDNA clones (33 per time point) and 287 sequences (median of 7 per time point). We observed a trend toward lower d(N)/d(S) after HIV seroconversion in 7 of 10 subjects and lower d(N)/d(S) in those with rapid HIV disease progression. However, the magnitude of these differences was small. These results are consistent with the hypothesis that HIV infection alters the HCV quasispecies, but the number of subjects and observation time may be too low to characterize the full effect.
Figures
FIG. 1
Hypothetical course of HIV-1 seroconverter to show study conventions. For example, pre-SC1 refers to the first serum specimen in which the HCV quasispecies was studied. Pre-HIV refers to sequence evolution before HIV-1 seroconversion, as measured at pre-SC1 and pre-SC2. HIV seroconversion was estimated as the midpoint between the last HIV antibody (Ab)-negative and first HIV antibody-positive specimens.
FIG. 2
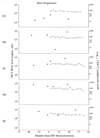
HCV RNA levels (ovals) and CD4 lymphocyte counts (squares) with respect to HIV seroconversion. Subjects are identified at the left as designated in Table 1. The four visits at which the HCV quasispecies was assessed correspond to HCV RNA determinations.
FIG. 2
HCV RNA levels (ovals) and CD4 lymphocyte counts (squares) with respect to HIV seroconversion. Subjects are identified at the left as designated in Table 1. The four visits at which the HCV quasispecies was assessed correspond to HCV RNA determinations.
FIG. 3
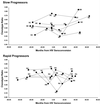
Clonotype ratios in 10 HIV seroconverters. Each point represents a unique subject visit. Slow and rapid progressors are identified by designations used in Table 1.
FIG. 4
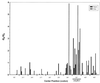
Overall dN/dS of envelope sequences at the amino acid positions in the HCV-1 polyprotein according to HIV progression status (5). Results were calculated comparing pre-SC1 with post-SC2 sequences using VarPlot configured with a 30-amino-acid sliding window centered on HVR1, as previously described (27). Similar plots were generated using a 10-amino-acid window and analysis of dN alone (not shown).
FIG. 5
Pre- and post-HIV dN/dS in rapid and slow progressors. (A) Sequences determined from cDNA clones representing at least 70% of 33 clones examined from each serum specimen, based on HDA+SSCP. (B) Sequences determined by direct sequencing of reverse transcription-PCR products. Relationship of pre-HIV and post-HIV to HIV seroconversion is as defined in the legend to Fig. 1.
Similar articles
- Multigene tracking of quasispecies in viral persistence and clearance of hepatitis C virus.
Chen S, Wang YM. Chen S, et al. World J Gastroenterol. 2005 May 21;11(19):2874-84. doi: 10.3748/wjg.v11.i19.2874. World J Gastroenterol. 2005. PMID: 15902722 Free PMC article. - Trends for genetic variation of Hepatitis C Virus quasispecies in Human Immunodeficiency virus-1 coinfected patients.
López-Labrador FX, Dove L, Hui CK, Phung Y, Kim M, Berenguer M, Wright TL. López-Labrador FX, et al. Virus Res. 2007 Dec;130(1-2):285-91. doi: 10.1016/j.virusres.2007.05.016. Epub 2007 Jun 29. Virus Res. 2007. PMID: 17601623 Free PMC article. - Evolution of HVR-1 quasispecies after 1-year treatment in HIV/HCV-coinfected patients according to the pattern of response to highly active antiretroviral therapy.
Solmone M, Girardi E, Lalle E, Abbate I, D'Arminio Monforte A, Cozzi-Lepri A, Alessandrini A, Piscopo R, Ebo F, Cosco L, Antonucci G, Ippolito G, Capobianchi MR; Hepal.Co.N.A. Study Group; I.Co.N.A Study Group. Solmone M, et al. Antivir Ther. 2006;11(1):87-94. Antivir Ther. 2006. PMID: 16518964 - HIV infection and antiretroviral therapy: effect on hepatitis C virus quasispecies variability.
Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, Thomassen LV, Emerson SS, Gretch DR. Shuhart MC, et al. J Infect Dis. 2006 May 1;193(9):1211-8. doi: 10.1086/502974. Epub 2006 Mar 28. J Infect Dis. 2006. PMID: 16586356 - Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus.
Toyoda H, Fukuda Y, Koyama Y, Takamatsu J, Saito H, Hayakawa T. Toyoda H, et al. J Hepatol. 1997 May;26(5):975-82. doi: 10.1016/s0168-8278(97)80105-5. J Hepatol. 1997. PMID: 9186827
Cited by
- Hepatitis C Virus (HCV) NS3 sequence diversity and antiviral resistance-associated variant frequency in HCV/HIV coinfection.
Jabara CB, Hu F, Mollan KR, Williford SE, Menezes P, Yang Y, Eron JJ, Fried MW, Hudgens MG, Jones CD, Swanstrom R, Lemon SM. Jabara CB, et al. Antimicrob Agents Chemother. 2014 Oct;58(10):6079-92. doi: 10.1128/AAC.03466-14. Epub 2014 Aug 4. Antimicrob Agents Chemother. 2014. PMID: 25092699 Free PMC article. - Evaluation of rapid progressors in HIV infection as an extreme phenotype.
Olson AD, Guiguet M, Zangerle R, Gill J, Perez-Hoyos S, Lodi S, Ghosn J, Dorrucci M, Johnson A, Sannes M, Moreno S, Porter K; for CASCADE Collaboration in EuroCoord. Olson AD, et al. J Acquir Immune Defic Syndr. 2014 Sep 1;67(1):15-21. doi: 10.1097/QAI.0000000000000240. J Acquir Immune Defic Syndr. 2014. PMID: 24872130 Free PMC article. - Evidence of distinct populations of hepatitis C virus in the liver and plasma of patients co-infected with HIV and HCV.
Blackard JT, Ma G, Sengupta S, Martin CM, Powell EA, Shata MT, Sherman KE. Blackard JT, et al. J Med Virol. 2014 Aug;86(8):1332-41. doi: 10.1002/jmv.23968. Epub 2014 Apr 30. J Med Virol. 2014. PMID: 24788693 Free PMC article. - Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses.
Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, Cox AL, Ray SC. Osburn WO, et al. Hepatology. 2014 Jun;59(6):2140-51. doi: 10.1002/hep.27013. Epub 2014 Apr 29. Hepatology. 2014. PMID: 24425349 Free PMC article. - Vertical transmission of hepatitis C virus: a tale of multiple outcomes.
Escobar-Gutiérrez A, Soudeyns H, Larouche A, Carpio-Pedroza JC, Martinez-Guarneros A, Vazquez-Chacon CA, Fonseca-Coronado S, Yamasaki LH, Ruiz-Tovar K, Cruz-Rivera M. Escobar-Gutiérrez A, et al. Infect Genet Evol. 2013 Dec;20:465-70. doi: 10.1016/j.meegid.2013.10.005. Epub 2013 Oct 18. Infect Genet Evol. 2013. PMID: 24140559 Free PMC article.
References
- Barnes W M. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. - PubMed
- Benhamou Y, Bochet M, Di M, Charlotte V F, Azria F, Coutellier A, Vidaud M, Bricaire F, Opolon P, Katlama C, Poynard T. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30:1054–1058. - PubMed
- Booth J C, Kumar U, Webster D, Monjardino J, Thomas H C. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology. 1998;27:223–227. - PubMed
- Centers for Disease Control and Prevention. 1999 USPHS/IDSA guidelines for the prevention of opportunistic infections in persons infected with human immunodeficiency virus: disease-specific recommendations. USPHS/IDSA Prevention of Opportunistic Infections Working Group. U.S. Public Health Services/Infectious Diseases Society of America. Morbid Mortal Wkly Rep. 1999;48:1–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DA004334/DA/NIDA NIH HHS/United States
- DA 12568/DA/NIDA NIH HHS/United States
- R56 DA004334/DA/NIDA NIH HHS/United States
- R01 DA012568/DA/NIDA NIH HHS/United States
- Z99 AI999999/Intramural NIH HHS/United States
- DA 04334/DA/NIDA NIH HHS/United States
- R56 DA012568/DA/NIDA NIH HHS/United States
- R37 DA004334/DA/NIDA NIH HHS/United States
- DA 10627/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases