Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast - PubMed (original) (raw)
Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast
D J Galgoczy et al. Mol Cell Biol. 2001 Mar.
Abstract
Despite the fact that eukaryotic cells enlist checkpoints to block cell cycle progression when their DNA is damaged, cells still undergo frequent genetic rearrangements, both spontaneously and in response to genotoxic agents. We and others have previously characterized a phenomenon (adaptation) in which yeast cells that are arrested at a DNA damage checkpoint eventually override this arrest and reenter the cell cycle, despite the fact that they have not repaired the DNA damage that elicited the arrest. Here, we use mutants that are defective in checkpoint adaptation to show that adaptation is important for achieving the highest possible viability after exposure to DNA-damaging agents, but it also acts as an entrée into some forms of genomic instability. Specifically, the spontaneous and X-ray-induced frequencies of chromosome loss, translocations, and a repair process called break-induced replication occur at significantly reduced rates in adaptation-defective mutants. This indicates that these events occur after a cell has first arrested at the checkpoint and then adapted to that arrest. Because malignant progression frequently involves loss of genes that function in DNA repair, adaptation may promote tumorigenesis by allowing genomic instability to occur in the absence of repair.
Figures
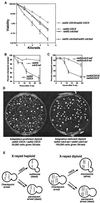
FIG. 1
Adaptation increases X-ray resistance in diploids. (A) Strains were plated and X-irradiated, and all resulting colonies were counted, regardless of their growth rate. Haploid strains (rad52 cdc5-ad or rad52 CDC5) were mated to themselves to produce isogenic diploids (20). (B and C) Cells were irradiated with 3 kilorads, and microcolony assays were performed to determine the percentage of cells that had adapted, as described elsewhere (20). (D) Either 30,000 cells (rad52 CDC5/rad52 CDC5 diploids, left) or 100,000 cells (rad52 cdc5-ad/rad52 cdc5-ad diploids, right) were plated, subjected to 3 kilorads of X-irradiation, and allowed to form colonies. (E) Model showing one broken and two unbroken chromosomes (each line represents two identical sisters) in haploid and diploid strains. Adaptation generates viable but karyotypically altered monosomic diploids.
FIG. 2
Adaptation precedes spontaneous chromosome loss. The frequency of spontaneous chromosome loss was determined using haploid strains harboring an extra copy of chromosome VII containing the CYH2 and ADE3 genes. Haploid CDC5 rad52 and cdc5-ad rad52 disomic strains (as in Table 1) were grown on nonselective plates and photographed. Chromosome losses appear as white sectors.
FIG. 3
Adaptation is required for spontaneous and damage-induced BIR events. Haploid CDC5 rad51 and cdc5-ad rad51 strains (as in Table 1) were altered to replace the centromere-linked LEU1 gene on the control chromosome with the URA3 gene (shown as B and b::URA3, respectively). Markers A, D, E, and F represent ADE3, ARO2, LYS5, and adh4::HIS3, respectively. Control chromosome DNA is shown in outline. For spontaneous events, strains were plated on medium lacking leucine (selecting for marker B) and containing cycloheximide (selecting against CYH2). The prominent classes of rearrangements are diagrammed. For X-ray induced events, cells were plated on nonselective plates, subjected to 3 kilorads of X rays, allowed to form colonies, and replica plated on cycloheximide plates lacking leucine to screen for BIR events.
FIG. 4
Translocations and BIR can both arise from a DNA lesion. (A) This diagram illustrates the expected outcome of a reciprocal recombination event in G2 between sister chromatids of different homologs. Segregation of sisters after mitosis will yield two possible outcomes: (i) both daughters can have markers identical to the starting strain (not shown), or (ii) each daughter can be homozygous for markers distal to the site of recombination (shown). (B) Outline of the experiment, the results of which are shown in panel C. Disomic rad51 CDC5 colonies (from irradiated nonselective plates) were scored for BIR events (as described in Materials and Methods). Colonies found to contain cells that had undergone BIR were restreaked from the original nonselective plate, and the resultant colonies were analyzed genetically for rearrangements of chromosome VII (BIR and/or deletions-translocations, as determined genetically). (C) DNA from the following strains were run on CHEF gels, blotted, and probed for the test chromosome: “starting strain” and “control strain” are marker strains with and without the test chromosome, respectively; 13 translocations-truncations isolated after restreaking 13 independent BIR-containing colonies (one translocation-truncation from each original colony) (lanes 1 to 13); and 4 translocations-truncations (lanes 14A to D) and 2 BIR (lanes 14 BIR E and F) colonies isolated from the same irradiated BIR-containing colony (6 total colonies isolated from one original colony). (D) Model showing three pairs of sister chromatids; the black and outlined sets represent the test and control chromosomes, respectively, and gray represents a different chromosome. Adaptation results in daughters that repair independently.
FIG. 5
A large region of chromosome VII flanked by two homologous tRNA genes is frequently deleted. (A) Marker strains with (lane 1) or without (lane 2) the control chromosome; 13 strains identified as large internal deletions from rad51 CDC5 (lanes 3 to 15) or rad51 cdc5-ad (lanes 16 to 28) were run on a CHEF gel, blotted, and probed for the test chromosome. (B) DNA from the starting strain or a strain containing a large internal deletion (as in panel A, lane 3) was fluorescently labeled and hybridized to a DNA array as described previously (9). Hybridization ratios are shown for genes on the left arm of chromosome VII. Arrows designate leucine tRNA genes flanking the deletion. (C) Oligonucleotides corresponding to the outside of the deletion were used in a PCR on the strains shown in panel A. M, marker. (D) The putative SSA intermediate between the centromere proximal [TL (CAA) G2] and distal [TL (CAA) G1] tRNA genes flanking the internal deletions mapped in panels A to C. The oligonucleotides used in panel C are indicated as arrows.
Similar articles
- Checkpoint adaptation in human cells.
Syljuåsen RG. Syljuåsen RG. Oncogene. 2007 Aug 30;26(40):5833-9. doi: 10.1038/sj.onc.1210402. Epub 2007 Mar 26. Oncogene. 2007. PMID: 17384683 Review. - Checkpoint adaptation in recombination-deficient cells drives aneuploidy and resistance to genotoxic agents.
Vydzhak O, Bender K, Klermund J, Busch A, Reimann S, Luke B. Vydzhak O, et al. DNA Repair (Amst). 2020 Nov;95:102939. doi: 10.1016/j.dnarep.2020.102939. Epub 2020 Jul 30. DNA Repair (Amst). 2020. PMID: 32777450 - Two alternative cell cycle checkpoint pathways differentially control DNA damage-dependent induction of MAG1 and DDI1 expression in yeast.
Zhu Y, Xiao W. Zhu Y, et al. Mol Genet Genomics. 2001 Nov;266(3):436-44. doi: 10.1007/s004380100538. Mol Genet Genomics. 2001. PMID: 11713673
Cited by
- Reduced kinase activity of polo kinase Cdc5 affects chromosome stability and DNA damage response in S. cerevisiae.
Rawal CC, Riccardo S, Pesenti C, Ferrari M, Marini F, Pellicioli A. Rawal CC, et al. Cell Cycle. 2016 Nov;15(21):2906-2919. doi: 10.1080/15384101.2016.1222338. Epub 2016 Aug 26. Cell Cycle. 2016. PMID: 27565373 Free PMC article. - Adaptation to DNA damage checkpoint in senescent telomerase-negative cells promotes genome instability.
Coutelier H, Xu Z, Morisse MC, Lhuillier-Akakpo M, Pelet S, Charvin G, Dubrana K, Teixeira MT. Coutelier H, et al. Genes Dev. 2018 Dec 1;32(23-24):1499-1513. doi: 10.1101/gad.318485.118. Epub 2018 Nov 21. Genes Dev. 2018. PMID: 30463903 Free PMC article. - Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway.
Donnianni RA, Ferrari M, Lazzaro F, Clerici M, Tamilselvan Nachimuthu B, Plevani P, Muzi-Falconi M, Pellicioli A. Donnianni RA, et al. PLoS Genet. 2010 Jan 22;6(1):e1000763. doi: 10.1371/journal.pgen.1000763. PLoS Genet. 2010. PMID: 20098491 Free PMC article. - The many types of heterogeneity in replicative senescence.
Xu Z, Teixeira MT. Xu Z, et al. Yeast. 2019 Nov;36(11):637-648. doi: 10.1002/yea.3433. Epub 2019 Aug 6. Yeast. 2019. PMID: 31306505 Free PMC article. Review. - Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination.
Delmas S, Shunburne L, Ngo HP, Allers T. Delmas S, et al. PLoS Genet. 2009 Jul;5(7):e1000552. doi: 10.1371/journal.pgen.1000552. Epub 2009 Jul 10. PLoS Genet. 2009. PMID: 19593371 Free PMC article.
References
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. . (Erratum, 282:1421.) - PubMed
- Featherstone C, Jackson S P. DNA double-strand break repair. Curr Biol. 1999;9:R759–R761. - PubMed
- Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases