Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control - PubMed (original) (raw)
Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control
D J Clarke et al. Mol Cell Biol. 2001 Mar.
Abstract
In budding yeast, anaphase initiation is controlled by ubiquitin-dependent degradation of Pds1p. Analysis of pds1 mutants implicated Pds1p in the DNA damage, spindle assembly, and S-phase checkpoints. Though some components of these pathways are known, others remain to be identified. Moreover, the essential function of Pds1p, independent of its role in checkpoint control, has not been elucidated. To identify loci that genetically interact with PDS1, we screened for dosage suppressors of a temperature-sensitive pds1 allele, pds1-128, defective for checkpoint control at the permissive temperature and essential for viability at 37 degrees C. Genetic and functional interactions of two suppressors are described. RAD23 and DDI1 suppress the temperature and hydroxyurea, but not radiation or nocodazole, sensitivity of pds1-128. rad23 and ddi1 mutants are partially defective in S-phase checkpoint control but are proficient in DNA damage and spindle assembly checkpoints. Therefore, Rad23p and Ddi1p participate in a subset of Pds1p-dependent cell cycle controls. Both Rad23p and Ddi1p contain ubiquitin-associated (UBA) domains which are required for dosage suppression of pds1-128. UBA domains are found in several proteins involved in ubiquitin-dependent proteolysis, though no function has been assigned to them. Deletion of the UBA domains of Rad23p and Ddi1p renders cells defective in S-phase checkpoint control, implicating UBA domains in checkpoint signaling. Since Pds1p destruction, and thus checkpoint regulation of mitosis, depends on ubiquitin-dependent proteolysis, we propose that the UBA domains functionally interact with the ubiquitin system to control Pds1p degradation in response to checkpoint activation.
Figures
FIG. 1
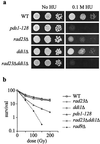
Dosage suppressors of pds1-128. (a) Suppression of pds1-128 but not pds1Δ temperature sensitivity by episomal plasmids maintained at high copy number in yeast cells carrying YEp24(URA3), YEp-PDS1, YEp-RAD23, YEp-DDI1, and YEp-ESP1. Patches were grown at 25°C on synthetic medium containing dextrose but without uracil to prevent loss of the YEp24-based plasmids. These patches were replicated on to YEP-dextrose plates and grown at 25 to 37°C for 2 days. (b) Genetic interactions between GAL1:RAD23 or GAL1:DDI1 and temperature-sensitive esp1 strains. Strains carrying esp1 alleles (esp1-N5 and esp1-B7 [13]) with or without RAD23 and DDI1 alleles under control of the inducible GAL1 promoter were grown at 30°C on YEP-dextrose plates, then replicated to YEP-galactose plates, and grown for 2 days at 30 to 35°C. (c) Suppression of pds1-128 mec1-1 temperature sensitivity by episomal plasmids maintained at high copy number in yeast cells carrying YEp-PDS1, YEp-RAD23, YEp-DDI1, YEp24, and YCp50, grown as for panel a.
FIG. 2
Suppression of pds1-128 S-phase checkpoint defects. (a) Suppression of pds1-128 HU sensitivity by RAD23 and DDI1 overexpression. Strains were grown to midlog phase, and then serial dilutions were spotted on to YEP-dextrose or YEP-galactose plates (to repress or induce GAL1:RAD23 and GAL1:DDI1) containing HU and grown for 2 to 3 days. (b) Overexpression of RAD23 increases Pds1-128p levels in HU-treated cells. Cells containing a GAL1:RAD23 gene and a hemagglutinin (HA) epitope-tagged pds1-128 allele were grown to midlog phase (minus HU) or accumulated in S-phase with 100 mM HU for 3 h (plus HU) in dextrose (−) or galactose (+) medium to repress or induce GAL1:RAD23. Cellular protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Pds1-128-p-HA was detected after Western blotting. Cdc28p levels served as a loading control. A light exposure and a darker exposure of the same blot are shown.
FIG. 3
Suppression of pds1-128 temperature and HU sensitivity depends on Rad23p and Ddi1p UBA domains. (a) Structural similarity between RAD23 and DDI1. Box diagrams show relative sizes of each ORF and the size and position of UBA, XPC, and UBL domains. C-terminal UBA domains (right) share 38% identity. The position of Ddi1p L426 is indicated in the alignment below. A hypothetical fission yeast protein (gb|Z69728 and gi|1204230) has an overall 32% identity to budding yeast Ddi1p. Probable Ddi1p homologues giving high identity scores are present in Homo sapiens (gb|AA406136 and gi|2064117; dbest database), Caenorhabditis elegans (gb|U50068 and gi|1208859) and Leishmania major (gb|AC002305 and gi|2429118). (b and c) Temperature sensitivity (b) and HU sensitivity (c) of pds1-128 strains overexpressing RAD23 and DDI1 or truncated versions of these genes expressing proteins lacking UBA domains or the RAD23 UBL. Similar results were obtained when the WT or truncated proteins were tested as (Rad23p-MYC6 and Ddi1p-His6) fusions. Strains were handled as described in the legends to Fig. 1b and 2a. (d) Lack of genetic interaction between GAL1:rad23Δuba2 and temperature-sensitive esp1. Strains were handled as described in the legend to Fig. 1b.
FIG. 4
rad23Δ ddi1Δ cells are HU sensitive but not radiation sensitive. (a) Sensitivity of pds1 and rad23Δ ddi1Δ mutants to HU. Strains were grown to midlog phase, then serial dilutions were spotted on to YEP-dextrose or YEP-dextrose plates containing HU, and growth continued for 2 to 3 days at 30°C. (b) Sensitivity of pds1, rad23Δ, ddi1Δ, and rad9Δ mutants to gamma irradiation. Strains were grown to midlog phase at 30°C in YEP-dextrose medium, irradiated, and then plated on YEP dextrose. Colonies were counted after 2 to 4 days.
FIG. 5
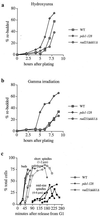
S-phase checkpoint defect of rad23Δ ddi1Δ mutants (a) Rebudding assay on HU plates. WT, pds1-128, and rad23Δ ddi1Δ cells were arrested in G1 at 30°C (α-factor), washed, and plated on YEP-dextrose plates containing 100 mM HU. Each strain budded with similar timing (not shown). Rebudding (the appearance of microcolonies containing three or more cell bodies) was scored at various time intervals (see also Fig. 6). (b) Rebudding assay following gamma irradiation. The procedure was as described for Fig. 5a. The ability of cells to rebud on YEP-dextrose plates following gamma irradiation in G1 was scored. (c) Timing of budding, G2 spindle formation, and spindle elongation in WT and mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor).
FIG. 6
S-phase checkpoint defect of rad23Δuba2 ddi1Δuba mutant cells. (a) Sensitivity of rad23Δuba2 ddi1Δuba mutant cells to HU. Strains were grown to midlog phase, then serial dilutions were spotted onto YEP-dextrose or YEP-dextrose plates containing HU, and growth continued for 2 to 3 days at 30°C. (b) Rebudding assay on HU plates or following gamma irradiation (see legend to Fig. 5 for details). Microcolonies were scored based on the number of cell bodies per microcolony. (c) Timing of short spindle formation, S-phase progression, and spindle elongation in WT and mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor). S-phase progression was monitored by FACScan analysis; histogram plots show DNA content throughout the experiment (in grey), and overlaid solid lines indicate positions of 1C and 2C DNA peaks taken from the asynchronous cultures prior to G1 arrest. The percentage of the total cells that had elongated spindles above each histogram. (d) Timing of loss of sister chromatid cohesion in WT and rad23Δuba2 ddi1Δuba mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor). Both strains budded and replicated DNA with similar timing (FACScan analysis not shown), but loss of cohesion occurred prematurely in the mutant cells. Cohesion was monitored at the TRP1 locus using the tetR-GFP/tetO system as previously described (4).
FIG. 7
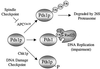
Model for control of Pds1p stability. Pds1p can become polyubiquitinated by APCCdc20, a multienzyme E3 ubiquitin ligase. The ubiquitin chain enables a 26S proteasome to recognize and degrade Pds1p. These events must occur prior to the onset of anaphase, since Pds1p inhibits the activity of separin Esp1p, needed for anaphase spindle elongation and loss of sister chromatid cohesion. To block anaphase onset, checkpoint controls prevent Pds1p degradation. The spindle assembly checkpoint directly inhibits APCCdc20, and the DNA damage checkpoint is thought to promote Pds1p stability by a Chk1p-dependent phosphorylation of Pds1p. We propose a novel mechanism for anaphase control, in which Rad23p and Ddi1p are able to recognize ubiquitinated Pds1p, or the APC (not shown), and thereby inhibit the ubiquitin chain elongation that is required for targeting Pds1p to the 26S proteasome.
Similar articles
- Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s).
Yamamoto A, Guacci V, Koshland D. Yamamoto A, et al. J Cell Biol. 1996 Apr;133(1):99-110. doi: 10.1083/jcb.133.1.99. J Cell Biol. 1996. PMID: 8601617 Free PMC article. - Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae.
Yamamoto A, Guacci V, Koshland D. Yamamoto A, et al. J Cell Biol. 1996 Apr;133(1):85-97. doi: 10.1083/jcb.133.1.85. J Cell Biol. 1996. PMID: 8601616 Free PMC article. - The Pds1 anaphase inhibitor and Mec1 kinase define distinct checkpoints coupling S phase with mitosis in budding yeast.
Clarke DJ, Segal M, Mondésert G, Reed SI. Clarke DJ, et al. Curr Biol. 1999 Apr 8;9(7):365-8. doi: 10.1016/s0960-9822(99)80163-8. Curr Biol. 1999. PMID: 10209118 - Cell cycle: cull and destroy.
Jackson PK. Jackson PK. Curr Biol. 1996 Oct 1;6(10):1209-12. doi: 10.1016/s0960-9822(96)00697-5. Curr Biol. 1996. PMID: 8939566 Review. - Regulation of S phase.
Dutta A. Dutta A. Results Probl Cell Differ. 1998;22:35-55. doi: 10.1007/978-3-540-69686-5_2. Results Probl Cell Differ. 1998. PMID: 9670318 Review. No abstract available.
Cited by
- Mammalian Ddi2 is a shuttling factor containing a retroviral protease domain that influences binding of ubiquitylated proteins and proteasomal degradation.
Collins GA, Sha Z, Kuo CL, Erbil B, Goldberg AL. Collins GA, et al. J Biol Chem. 2022 May;298(5):101875. doi: 10.1016/j.jbc.2022.101875. Epub 2022 Mar 28. J Biol Chem. 2022. PMID: 35358511 Free PMC article. - Proteasome condensate formation is driven by multivalent interactions with shuttle factors and ubiquitin chains.
Waite KA, Vontz G, Lee SY, Roelofs J. Waite KA, et al. Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2310756121. doi: 10.1073/pnas.2310756121. Epub 2024 Feb 26. Proc Natl Acad Sci U S A. 2024. PMID: 38408252 Free PMC article. - TaUBA, a UBA domain-containing protein in wheat (Triticum aestivum L.), is a negative regulator of salt and drought stress response in transgenic Arabidopsis.
Li X, Zhang SS, Ma JX, Guo GY, Zhang XY, Liu X, Bi CL. Li X, et al. Plant Cell Rep. 2015 May;34(5):755-66. doi: 10.1007/s00299-015-1739-3. Epub 2015 Jan 21. Plant Cell Rep. 2015. PMID: 25604990 - Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway.
Ortolan TG, Chen L, Tongaonkar P, Madura K. Ortolan TG, et al. Nucleic Acids Res. 2004 Dec 15;32(22):6490-500. doi: 10.1093/nar/gkh987. Print 2004. Nucleic Acids Res. 2004. PMID: 15601997 Free PMC article. - Human DNA-Damage-Inducible 2 Protein Is Structurally and Functionally Distinct from Its Yeast Ortholog.
Sivá M, Svoboda M, Veverka V, Trempe JF, Hofmann K, Kožíšek M, Hexnerová R, Sedlák F, Belza J, Brynda J, Šácha P, Hubálek M, Starková J, Flaisigová I, Konvalinka J, Šašková KG. Sivá M, et al. Sci Rep. 2016 Jul 27;6:30443. doi: 10.1038/srep30443. Sci Rep. 2016. PMID: 27461074 Free PMC article.
References
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. - PubMed
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
- Clarke D J, Segal M, Mondesert G, Reed S I. The Pds1 anaphase inhibitor and Mec1 kinase define distinct checkpoints coupling S phase with mitosis in budding yeast. Curr Biol. 1999;9:365–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases