P(II) signal transduction proteins, pivotal players in microbial nitrogen control - PubMed (original) (raw)
Review
P(II) signal transduction proteins, pivotal players in microbial nitrogen control
T Arcondéguy et al. Microbiol Mol Biol Rev. 2001 Mar.
Abstract
The P(II) family of signal transduction proteins are among the most widely distributed signal proteins in the bacterial world. First identified in 1969 as a component of the glutamine synthetase regulatory apparatus, P(II) proteins have since been recognized as playing a pivotal role in control of prokaryotic nitrogen metabolism. More recently, members of the family have been found in higher plants, where they also potentially play a role in nitrogen control. The P(II) proteins can function in the regulation of both gene transcription, by modulating the activity of regulatory proteins, and the catalytic activity of enzymes involved in nitrogen metabolism. There is also emerging evidence that they may regulate the activity of proteins required for transport of nitrogen compounds into the cell. In this review we discuss the history of the P(II) proteins, their structures and biochemistry, and their distribution and functions in prokaryotes. We survey data emerging from bacterial genome sequences and consider other likely or potential targets for control by P(II) proteins.
Figures
FIG. 1
Nitrogen regulation (Ntr) system of enteric bacteria. The activities of both GS and NtrC are regulated in response to the intracellular nitrogen status. UTase (glnD product) catalyzes the uridylylation and deuridylylation of PII(glnB product). ATase catalyzes the adenylylation and deadenylylation of GS. NtrB catalyzes the phosphorylation and dephosphorylation of NtrC.
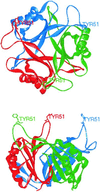
FIG. 2
Structure of the E. coli GlnB trimer viewed (top) from above and (bottom) from the side. Individual monomers are colored red, green, and blue.
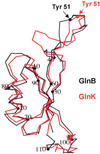
FIG. 3
Comparison of the structures of E. coli GlnB and GlnK. Superimposition of the Cα traces for GlnB (black) and GlnK (red) with the position of the Tyr-51 residues in each molecule is indicated. Adapted from reference .
FIG. 4
Schematic representation of the potential mutiplicity of roles for PII proteins. This cartoon summarizes information (both definitive and predicted) obtained from many different organisms.
Similar articles
- Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942.
Forchhammer K, Tandeau de Marsac N. Forchhammer K, et al. J Bacteriol. 1995 Apr;177(8):2033-40. doi: 10.1128/jb.177.8.2033-2040.1995. J Bacteriol. 1995. PMID: 7721695 Free PMC article. - PII, the key regulator of nitrogen metabolism in the cyanobacteria.
Zhang Y, Zhao J. Zhang Y, et al. Sci China C Life Sci. 2008 Dec;51(12):1056-65. doi: 10.1007/s11427-008-0148-z. Epub 2008 Dec 18. Sci China C Life Sci. 2008. PMID: 19093078 Review. - P(II) signal transduction proteins: nitrogen regulation and beyond.
Huergo LF, Chandra G, Merrick M. Huergo LF, et al. FEMS Microbiol Rev. 2013 Mar;37(2):251-83. doi: 10.1111/j.1574-6976.2012.00351.x. Epub 2012 Aug 28. FEMS Microbiol Rev. 2013. PMID: 22861350 Review. - Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli.
Atkinson MR, Ninfa AJ. Atkinson MR, et al. Mol Microbiol. 1998 Jul;29(2):431-47. doi: 10.1046/j.1365-2958.1998.00932.x. Mol Microbiol. 1998. PMID: 9720863 - PII signal transduction proteins.
Ninfa AJ, Atkinson MR. Ninfa AJ, et al. Trends Microbiol. 2000 Apr;8(4):172-9. doi: 10.1016/s0966-842x(00)01709-1. Trends Microbiol. 2000. PMID: 10754576 Review.
Cited by
- Specific gene responses of Rhodococcus jostii RHA1 during growth in soil.
Iino T, Wang Y, Miyauchi K, Kasai D, Masai E, Fujii T, Ogawa N, Fukuda M. Iino T, et al. Appl Environ Microbiol. 2012 Oct;78(19):6954-62. doi: 10.1128/AEM.00164-12. Epub 2012 Jul 27. Appl Environ Microbiol. 2012. PMID: 22843521 Free PMC article. - Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source.
Lie TJ, Leigh JA. Lie TJ, et al. J Bacteriol. 2002 Oct;184(19):5301-6. doi: 10.1128/JB.184.19.5301-5306.2002. J Bacteriol. 2002. PMID: 12218015 Free PMC article. - Effect of perturbation of ATP level on the activity and regulation of nitrogenase in Rhodospirillum rubrum.
Zhang Y, Pohlmann EL, Roberts GP. Zhang Y, et al. J Bacteriol. 2009 Sep;191(17):5526-37. doi: 10.1128/JB.00585-09. Epub 2009 Jun 19. J Bacteriol. 2009. PMID: 19542280 Free PMC article. - Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations.
Levicán G, Ugalde JA, Ehrenfeld N, Maass A, Parada P. Levicán G, et al. BMC Genomics. 2008 Dec 3;9:581. doi: 10.1186/1471-2164-9-581. BMC Genomics. 2008. PMID: 19055775 Free PMC article. - Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1.
Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM. Gutiérrez RA, et al. Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4939-44. doi: 10.1073/pnas.0800211105. Epub 2008 Mar 14. Proc Natl Acad Sci U S A. 2008. PMID: 18344319 Free PMC article.
References
- Adler S P, Purich D, Stadtman E R. Cascade control of Escherichia coli glutamine synthetase: properties of the PIIregulatory protein and the uridylyltransferase-uridylylremoving enzyme. J Biol Chem. 1975;250:6264–6272. - PubMed
- Allikmets R, Gerrard B, Court D, Dean M. Cloning and organisation of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene. 1993;136:231–236. - PubMed
- Amar M, Patriarca E J, Manco G, Bernard P, Riccio A, Lamberti A, Defez R, Iaccarino M. Regulation of nitrogen metabolism is altered in a glnB mutant strain of Rhizobium leguminosarum. Mol Microbiol. 1994;11:685–693. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources