Gene transfer to the desiccation-tolerant cyanobacterium Chroococcidiopsis - PubMed (original) (raw)
Gene transfer to the desiccation-tolerant cyanobacterium Chroococcidiopsis
D Billi et al. J Bacteriol. 2001 Apr.
Abstract
The coccoid cyanobacterium Chroococcidiopsis dominates microbial communities in the most extreme arid hot and cold deserts. These communities withstand constraints that result from multiple cycles of drying and wetting and/or prolonged desiccation, through mechanisms which remain poorly understood. Here we describe the first system for genetic manipulation of Chroococcidiopsis. Plasmids pDUCA7 and pRL489, based on the pDU1 replicon of Nostoc sp. strain PCC 7524, were transferred to different isolates of Chroococcidiopsis via conjugation and electroporation. This report provides the first evidence that pDU1 replicons can be maintained in cyanobacteria other than Nostoc and Anabaena. Following conjugation, both plasmids replicated in Chroococcidiopsis sp. strains 029, 057, and 123 but not in strains 171 and 584. Both plasmids were electroporated into strains 029 and 123 but not into strains 057, 171, and 584. Expression of P(psbA)-luxAB on pRL489 was visualized through in vivo luminescence. Efficiencies of conjugative transfer for pDUCA7 and pRL489 into Chroococcidiopsis sp. strain 029 were approximately 10(-2) and 10(-4) transconjugants per recipient cell, respectively. Conjugative transfer occurred with a lower efficiency into strains 057 and 123. Electrotransformation efficiencies of about 10(-4) electrotransformants per recipient cell were achieved with strains 029 and 123, using either pDUCA7 or pRL489. Extracellular deoxyribonucleases were associated with each of the five strains. Phylogenetic analysis, based upon the V6 to V8 variable regions of 16S rRNA, suggests that desert strains 057, 123, 171, and 029 are distinct from the type species strain Chroococcidiopsis thermalis PCC 7203. The high efficiency of conjugative transfer of Chroococcidiopsis sp. strain 029, from the Negev Desert, Israel, makes this a suitable experimental strain for genetic studies on desiccation tolerance.
Figures
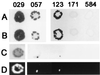
FIG. 1
Typical result of spot matings using Chroococcidiopsis sp. strains 029, 057, 123, 171, and 584. Rows: A, mobilization of plasmid pDUCA7 using conjugative plasmid pK2013 in the presence of helper plasmid pRL528; B, mobilization of plasmid pDUCA7 using conjugative plasmid pK2013 in the absence of helper plasmid pRL528; C, presumptive transconjugants of Chroococcidiopsis cells bearing plasmid pRL489 obtained using conjugative plasmid pRL443 and helper plasmid pRL528; D, luminescence was observed in colonies (see Fig. 1C) obtained from strains 029, 057, and 123 and not in the areas where strains 171 and 584 were used as the recipients. Filters shown were collected after 30 days of incubation.
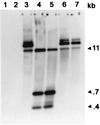
FIG. 2
Southern analysis of Chroococcidiopsis sp. strain 029 transconjugants (isolate CH91B1) obtained after mobilization of pDUCA7. The probe was labelled pDUCA7. Lanes: 1, 1-kb DNA ladder (LTI); 2, total DNA from Chroococcidiopsis sp. strain 029 (wild type); 3, total DNA from Chroococcidiopsis strain 029 isolate CH91B1; 4, total DNA from Chroococcidiopsis strain 029 isolate CH91B1 digested with _Pst_I; 5, authentic pDUCA7 digested with _Pst_I; 6, undigested pDUCA7 from E. coli DH10B transformed with plasmid DNA extracted from Chroococcidiopsis sp. strain 029 isolate CH91B1; 7, authentic pDUCA7.
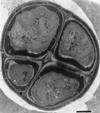
FIG. 3
Electron micrograph of ultrathin section of 2-month-old Chroococcidiopsis strain 029 showing multicellular aggregate containing cells of different sizes. An electron-translucent layer (white) and a fibrillar electron-dense envelope of capsular polysaccharide (grey) surround the aggregate. Bar = 0.5 μm.
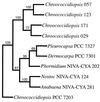
FIG. 4
Phylogenetic analysis of partial 16S rDNA. An unrooted consensus tree is shown. The numbers at the tree forks indicate the number of times out of 100 data sets that the strains to the right of the fork clustered (only numbers greater than 50 are shown).
Similar articles
- Avoidance of protein oxidation correlates with the desiccation and radiation resistance of hot and cold desert strains of the cyanobacterium Chroococcidiopsis.
Fagliarone C, Mosca C, Ubaldi I, Verseux C, Baqué M, Wilmotte A, Billi D. Fagliarone C, et al. Extremophiles. 2017 Nov;21(6):981-991. doi: 10.1007/s00792-017-0957-8. Epub 2017 Aug 30. Extremophiles. 2017. PMID: 28856526 - Plasmid stability in dried cells of the desert cyanobacterium Chroococcidiopsis and its potential for GFP imaging of survivors on Earth and in space.
Billi D. Billi D. Orig Life Evol Biosph. 2012 Jun;42(2-3):235-45. doi: 10.1007/s11084-012-9277-2. Epub 2012 May 26. Orig Life Evol Biosph. 2012. PMID: 22638838 - Salt tolerance and polyphyly in the cyanobacterium Chroococcidiopsis (Pleurocapsales).
Cumbers J, Rothschild LJ. Cumbers J, et al. J Phycol. 2014 Jun;50(3):472-82. doi: 10.1111/jpy.12169. Epub 2014 Mar 15. J Phycol. 2014. PMID: 26988320 - Mutations stabilize small subunit ribosomal RNA in desiccation-tolerant cyanobacteria nostoc.
Han D, Hu Z. Han D, et al. Curr Microbiol. 2007 Apr;54(4):254-9. doi: 10.1007/s00284-006-0095-5. Epub 2007 Mar 2. Curr Microbiol. 2007. PMID: 17334839 - Identification of the region of cyanobacterial plasmid pDU1 necessary for replication in Anabaena sp. strain M-131.
Schmetterer G, Wolk CP. Schmetterer G, et al. Gene. 1988;62(1):101-9. doi: 10.1016/0378-1119(88)90583-5. Gene. 1988. PMID: 3131190
Cited by
- First characterization of cultivable extremophile Chroococcidiopsis isolates from a solar panel.
Baldanta S, Arnal R, Blanco-Rivero A, Guevara G, Navarro Llorens JM. Baldanta S, et al. Front Microbiol. 2023 Feb 17;14:982422. doi: 10.3389/fmicb.2023.982422. eCollection 2023. Front Microbiol. 2023. PMID: 36876112 Free PMC article. - Gloeocapsopsis AAB1, an extremely desiccation-tolerant cyanobacterium isolated from the Atacama Desert.
Azua-Bustos A, Zúñiga J, Arenas-Fajardo C, Orellana M, Salas L, Rafael V. Azua-Bustos A, et al. Extremophiles. 2014 Jan;18(1):61-74. doi: 10.1007/s00792-013-0592-y. Epub 2013 Oct 20. Extremophiles. 2014. PMID: 24141552 - Assessing horizontal gene transfer in the rhizosphere of Brachypodium distachyon using fabricated ecosystems (EcoFABs).
Priya S, Rossbach S, Eng T, Lin H-H, Andeer PF, Mortimer JC, Northen TR, Mukhopadhyay A. Priya S, et al. Appl Environ Microbiol. 2024 Nov 20;90(11):e0150524. doi: 10.1128/aem.01505-24. Epub 2024 Nov 4. Appl Environ Microbiol. 2024. PMID: 39494898 Free PMC article. - Gene transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts.
Taton A, Lis E, Adin DM, Dong G, Cookson S, Kay SA, Golden SS, Golden JW. Taton A, et al. PLoS One. 2012;7(1):e30901. doi: 10.1371/journal.pone.0030901. Epub 2012 Jan 24. PLoS One. 2012. PMID: 22292073 Free PMC article. - Bacterial life and dinitrogen fixation at a gypsum rock.
Boison G, Mergel A, Jolkver H, Bothe H. Boison G, et al. Appl Environ Microbiol. 2004 Dec;70(12):7070-7. doi: 10.1128/AEM.70.12.7070-7077.2004. Appl Environ Microbiol. 2004. PMID: 15574902 Free PMC article.
References
- Billi D, Potts M. Life without water: responses of prokaryotes to desiccation. In: Storey K B, Storey J M, editors. Environmental stressors and gene responses. Amsterdam, The Netherlands: Elsevier; 2000. pp. 181–192.
- Cohen M F, Meeks J C, Cai Y A, Wolk C P. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 1998;297:3–17.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources