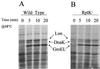Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B) - PubMed (original) (raw)
Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B)
S Zhang et al. J Bacteriol. 2001 Apr.
Abstract
sigma(B), the general stress response sigma factor of Bacillus subtilis, is activated when the cell's energy levels decline or the bacterium is exposed to environmental stress (e.g., heat shock, ethanol). Physical stress activates sigma(B) through a collection of regulatory kinases and phosphatases (the Rsb proteins) which catalyze the release of sigma(B) from an anti-sigma(B) factor inhibitor. The means by which diverse stresses communicate with the Rsb proteins is unknown; however, a role for the ribosome in this process was suggested when several of the upstream members of the sigma(B) stress activation cascade (RsbR, -S, and -T) were found to cofractionate with ribosomes in crude B. subtilis extracts. We now present evidence for the involvement of a ribosome-mediated process in the stress activation of sigma(B). B. subtilis strains resistant to the antibiotic thiostrepton, due to the loss of ribosomal protein L11 (RplK), were found to be blocked in the stress activation of sigma(B). Neither the energy-responsive activation of sigma(B) nor stress-dependent chaperone gene induction (a sigma(B)-independent stress response) was inhibited by the loss of L11. The Rsb proteins required for stress activation of sigma(B) are shown to be active in the RplK(-) strain but fail to be triggered by stress. The data demonstrate that the B. subtilis ribosomes provide an essential input for the stress activation of sigma(B) and suggest that the ribosomes may themselves be the sensors for stress in this system.
Figures
FIG. 1
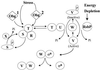
Activation of ςB. ςB is held inactive in unstressed B. subtilis as a complex with an anti-ςB protein, RsbW (W). ςB is freed from RsbW when a release factor, RsbV (V), binds to RsbW. In unstressed B. subtilis, RsbV is inactive due to an RsbW-catalyzed phosphorylation (V-P). Environmental stress activates an RsbV-P phosphatase, RsbU (U), which reactivates RsbV. RsbT (T) is the RsbU activator. RsbT is normally bound to a negative regulator, RsbS (S), which inhibits its activity. RsbR (R) also binds to RsbS and -T and is believed to facilitate their interactions. Upon exposure to stress, RsbT phosphorylates and inactivates RsbS and then activates the RsbU phosphatase. Obg, an essential GTPase that can also bind to RsbT, is required for stress to trigger the activation of RsbT. It is unknown whether an Obg-dependent process serves as a coinductant for stress to activate RsbT (process 1) or as the vehicle through which stress directly communicates to RsbT (process 2). RsbS-P is dephosphorylated and reactivated by a phosphatase, RsbX (X), that is encoded by one of the genes downstream of the sigB operon's ςB-dependent promoter. RsbX levels become elevated when ςB is active, which may facilitate a return of RsbT to an inactive complex with RsbS. Energy depletion activates a separate pathway in which a novel RsbV-P phosphatase (RsbP) is triggered, by unknown means, to reactivate RsbV. This model is based on references , , , , , , , , , and .
FIG. 2
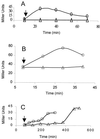
Activation of ςB by ethanol stress or sodium azide. B. subtilis strains growing in LB were treated with either 4% ethanol or 2 mM sodium azide. Culture samples were taken at the indicated times and analyzed for ςB-dependent β-galactosidase activity. The arrows indicate the times at which either ethanol or azide was added to the cultures. (A) BSA46 (○) wild-type and BSZ9 (▵) RplK− strains exposed to ethanol at an _A_540 of approximately 0.2. (B) Strain BSZ5 (RelA−) treated with ethanol at an _A_540 of 0.1 (○) or untreated (▵). (C) BSA46 (○) wild-type and BSZ9 (▵) RplK− strains treated with azide at an _A_540 of 0.35.
FIG. 3
Heat shock induction of chaperone proteins in RplK+ and RplK− B. subtilis strains. B. subtilis strains BSA46 (A) and BSZ9 rplK57 (B) were grown to an _A_540 of 0.3 and pulse-labeled for 5 min with 35[S]Met-Cys (1 μCi/ml) either at 37°C (0) or at 5, 10, or 20 min after transfer to 48°C (33). Cell lysates were fractionated by SDS-PAGE, and labeled protein bands were visualized by fluorography. The positions to which B. subtilis proteins with the molecular weights of Lon, DnaK, and GroEL would migrate in our gel system are indicated (6).
FIG. 4
Stress-dependent dephosphorylation of RsbV-P. BSA46 (wild-type) and BSZ9 (RplK−) cultures were grown in LB and exposed to ethanol (4% final concentration) during exponential growth at an _A_540 of 0.4. Bacteria were harvested before (0) and at various intervals after ethanol addition (2.5 to 10 min). Crude extracts were subjected to IEF and transferred to nitrocellulose, and the membrane was probed with an anti-RsbV monoclonal antibody (35). The positions to which phosphorylated (RsbV-P) and unphosphorylated RsbV migrate in this system are indicated.
FIG. 5
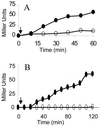
Activation of ςB by RsbT overexpression. Strains BSA419 (P_SPAC_::rsbT) (A) and BSZ10 (P_SPAC_::rsbT rplK57) (B) were grown in LB. At the times indicated by the arrows (_A_540, approximately 0.1), IPTG (1 mM) was added to half of each culture. Samples of the IPTG-induced (●) and control (○) cultures were analyzed for ςB-dependent β-galactosidase activity.
Similar articles
- Associations between Bacillus subtilis sigmaB regulators in cell extracts.
Kuo S, Zhang S, Woodbury RL, Haldenwang WG. Kuo S, et al. Microbiology (Reading). 2004 Dec;150(Pt 12):4125-36. doi: 10.1099/mic.0.27421-0. Microbiology (Reading). 2004. PMID: 15583165 - The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes.
Scott JM, Ju J, Mitchell T, Haldenwang WG. Scott JM, et al. J Bacteriol. 2000 May;182(10):2771-7. doi: 10.1128/JB.182.10.2771-2777.2000. J Bacteriol. 2000. PMID: 10781545 Free PMC article. - Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor sigma(B).
Scott JM, Haldenwang WG. Scott JM, et al. J Bacteriol. 1999 Aug;181(15):4653-60. doi: 10.1128/JB.181.15.4653-4660.1999. J Bacteriol. 1999. PMID: 10419966 Free PMC article. - The RsbRST stress module in bacteria: a signalling system that may interact with different output modules.
Pané-Farré J, Lewis RJ, Stülke J. Pané-Farré J, et al. J Mol Microbiol Biotechnol. 2005;9(2):65-76. doi: 10.1159/000088837. J Mol Microbiol Biotechnol. 2005. PMID: 16319496 Review. - Anti-sigma factor-mediated cell surface stress responses in Bacillus subtilis.
Asai K. Asai K. Genes Genet Syst. 2018 Apr 10;92(5):223-234. doi: 10.1266/ggs.17-00046. Epub 2018 Jan 17. Genes Genet Syst. 2018. PMID: 29343670 Review.
Cited by
- Novel roles of the master transcription factors Spo0A and sigmaB for survival and sporulation of Bacillus subtilis at low growth temperature.
Méndez MB, Orsaria LM, Philippe V, Pedrido ME, Grau RR. Méndez MB, et al. J Bacteriol. 2004 Feb;186(4):989-1000. doi: 10.1128/JB.186.4.989-1000.2004. J Bacteriol. 2004. PMID: 14761993 Free PMC article. - The universally conserved prokaryotic GTPases.
Verstraeten N, Fauvart M, Versées W, Michiels J. Verstraeten N, et al. Microbiol Mol Biol Rev. 2011 Sep;75(3):507-42, second and third pages of table of contents. doi: 10.1128/MMBR.00009-11. Microbiol Mol Biol Rev. 2011. PMID: 21885683 Free PMC article. Review. - Role of extracytoplasmic function sigma factor PG1660 (RpoE) in the oxidative stress resistance regulatory network of Porphyromonas gingivalis.
Dou Y, Rutanhira H, Chen X, Mishra A, Wang C, Fletcher HM. Dou Y, et al. Mol Oral Microbiol. 2018 Feb;33(1):89-104. doi: 10.1111/omi.12204. Epub 2017 Dec 15. Mol Oral Microbiol. 2018. PMID: 29059500 Free PMC article. - (p)ppGpp - an important player during heat shock response.
Driller K, Cornejo FA, Turgay K. Driller K, et al. Microlife. 2023 Apr 6;4:uqad017. doi: 10.1093/femsml/uqad017. eCollection 2023. Microlife. 2023. PMID: 37251512 Free PMC article. Review. - Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. Chaudhuri RR, et al. BMC Genomics. 2009 Jul 1;10:291. doi: 10.1186/1471-2164-10-291. BMC Genomics. 2009. PMID: 19570206 Free PMC article.
References
- Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. - PubMed
- Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases