Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma - PubMed (original) (raw)
Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma
C J DaFonseca et al. Proc Natl Acad Sci U S A. 2001.
Abstract
In response to IFN-gamma, the latent cytoplasmic Stat1 (signal transducer and activator of transcription) proteins translocate into the nucleus and activate transcription. We showed previously that Stat1 recruits a group of nuclear proteins, among them MCM5 (minichromosome maintenance) and MCM3, for transcription activation. MCM5 directly interacts with the transcription activation domain (TAD) of Stat1 and enhances Stat1-mediated transcription activation. In this report, we identified two specific residues (R732, K734) in MCM5 that are required for the direct interaction between Stat1 and MCM5 both in vitro and in vivo. MCM5 containing mutations of R732/K734 did not enhance Stat1-mediated transcription activation in response to IFN-gamma. In addition, it also failed to form complexes with other MCM proteins in vivo, suggesting that these two residues may be important for an interaction domain in MCM5. Furthermore, MCM5 bearing mutations in its ATPase and helicase domains did not enhance Stat1 activity. In vitro binding assays indicate that MCM3 does not interact directly with Stat1, suggesting that the presence of MCM3 in the group of Stat1TAD-interacting proteins is due to the association of MCM3 with MCM5. Finally, gel filtration analyses of nuclear extracts from INF-gamma-treated cells demonstrate that there is a MCM5/3 subcomplex coeluting with Stat1. Together, these results strongly suggest that Stat1 recruits a MCM5/3 subcomplex through direct interaction with MCM5 in the process of IFN-gamma-induced gene activation.
Figures
Figure 1
GST-Stat1TAD interacts directly with MCM5, but not MCM3. (A) Schematic view of Stat1α and -β and MCM5 and MCM3. The domain structure of Stat1 is according to Chen et al. (7). TAD, transcription activation domain; Y, Tyr-701; S, Ser-727. (B) MCM5 and MCM3 are among the group of Stat1TAD-interacting nuclear proteins. Nuclear extracts from U3A cells were incubated with Sepharose-bead-bound GST or GST-Stat1TAD fusion proteins. The bound proteins were separated by SDS/PAGE and analyzed by Western blotting with indicated antibodies. (C) Stat1TAD interacts directly with MCM5, but not with MCM3.35S-labeled MCM5 or MCM3 proteins were translated in vitro and incubated with the indicated GST fusion proteins bound to Sepharose beads. The bound proteins were separated by SDS/PAGE and visualized by autoradiography. The bottom panel shows the various GST fusion proteins separated by SDS/PAGE and visualized by Coomassie staining.
Figure 2
MCM5 protein containing mutations of R732 and K734 cannot interact with Stat1TAD. (A) Point mutations in MCM5./indicates a double mutation of residues; KMDA4 is a quintuplet mutation of the ATP-binding site K387 into Met and the conserved DEFD motif (445) into four Ala. Lines on top of the schematic MCM5 molecule indicate previously reported regions of MCM5 that were required for binding to Stat1TAD, with the thickness of the lines representing the strength of interaction. (B) Wild-type or mutant MCM5 proteins were labeled with 35S by in vitro translation and incubated with Sepharose-bead-bound GST or GST-Stat1TAD fusion proteins (GSTS1C). The proteins bound to beads were separated by SDS/PAGE and visualized by autoradiography. Input lanes contain 10% of total input.
Figure 3
The R732/K734 mutant MCM5 does not interact with Stat1 and other MCM proteins in vivo. (A) Wild-type or mutant MCM5 was tagged at the N terminus with the HA epitope and transiently transfected into 2fTGH cells. Stat1 proteins from whole-cell extracts were precipitated with a Stat1 antibody, and the immune precipitates were separated by SDS/PAGE and analyzed by Western blotting with the indicated antibodies. Input lanes contain 10% of total input. (B) Nuclear extracts from stable cell lines containing the various HA-tagged MCM5 proteins were prepared from cells treated with IFN-γ for 30 min. The HA-tagged MCM5 proteins were precipitated with the anti-HA antibody, and the immune precipitates were separated by SDS/PAGE and analyzed by Western blotting with the indicated antibodies. Input lanes contain 20% of total input.
Figure 4
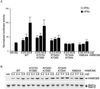
Specific interaction between Stat1 and MCM5 is required for MCM5 to enhance Stat1-mediated transcription activation. (A) Expression plasmids containing HA-tagged wild-type or mutant MCM5 were transiently transfected into U-2 OS cells together with a Stat1-dependent luciferase reporter and the internal control Renilla luciferase reporter (dual luciferase reporter system; Promega). Twenty-four hours after transfection, the cells were either left untreated or treated with IFN-γ for 6 h and harvested for luciferase assays. Results shown are luciferase activities normalized against internal control and the mean + SD of three to five experiments. For MCM5 overexpression samples, only results from treated cells are shown. Vec, the RcCMV plasmid. (B) Western blotting analyses were done with 10 μl of cell lysates used for luciferase assays above. Only lysates from treated cells are shown.
Figure 5
Coelution of Stat1 and the MCM5/3 subcomplex. Nuclear extracts from IFN-γ-treated (30 min) BUD-8 cells were fractionated by FPLC on a Superose 6 HR10/30 column. 2.5% of total input and 25% of each fraction (fraction numbers are at the top of each lane) were analyzed by Western blotting with the indicated antibodies. Molecular masses were calculated from a calibration curve generated with the Pharmacia HMW calibration kit.
Similar articles
- Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation.
Zhang JJ, Zhao Y, Chait BT, Lathem WW, Ritzi M, Knippers R, Darnell JE Jr. Zhang JJ, et al. EMBO J. 1998 Dec 1;17(23):6963-71. doi: 10.1093/emboj/17.23.6963. EMBO J. 1998. PMID: 9843502 Free PMC article. - The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation.
Snyder M, He W, Zhang JJ. Snyder M, et al. Proc Natl Acad Sci U S A. 2005 Oct 11;102(41):14539-44. doi: 10.1073/pnas.0507479102. Epub 2005 Sep 30. Proc Natl Acad Sci U S A. 2005. PMID: 16199513 Free PMC article. - The linker domain of Stat1 is required for gamma interferon-driven transcription.
Yang E, Wen Z, Haspel RL, Zhang JJ, Darnell JE Jr. Yang E, et al. Mol Cell Biol. 1999 Jul;19(7):5106-12. doi: 10.1128/MCB.19.7.5106. Mol Cell Biol. 1999. PMID: 10373559 Free PMC article. - Requirement of Ca2+ and CaMKII for Stat1 Ser-727 phosphorylation in response to IFN-gamma.
Nair JS, DaFonseca CJ, Tjernberg A, Sun W, Darnell JE Jr, Chait BT, Zhang JJ. Nair JS, et al. Proc Natl Acad Sci U S A. 2002 Apr 30;99(9):5971-6. doi: 10.1073/pnas.052159099. Epub 2002 Apr 23. Proc Natl Acad Sci U S A. 2002. PMID: 11972023 Free PMC article. - Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes.
Ouchi T, Lee SW, Ouchi M, Aaronson SA, Horvath CM. Ouchi T, et al. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5208-13. doi: 10.1073/pnas.080469697. Proc Natl Acad Sci U S A. 2000. PMID: 10792030 Free PMC article.
Cited by
- Expression of Minichromosome Maintenance Proteins (MCM) and Cancer Prognosis: A meta-analysis.
Gou K, Liu J, Feng X, Li H, Yuan Y, Xing C. Gou K, et al. J Cancer. 2018 Apr 6;9(8):1518-1526. doi: 10.7150/jca.22691. eCollection 2018. J Cancer. 2018. PMID: 29721062 Free PMC article. - Assembly of a parts list of the human mitotic cell cycle machinery.
Giotti B, Chen SH, Barnett MW, Regan T, Ly T, Wiemann S, Hume DA, Freeman TC. Giotti B, et al. J Mol Cell Biol. 2019 Aug 19;11(8):703-718. doi: 10.1093/jmcb/mjy063. J Mol Cell Biol. 2019. PMID: 30452682 Free PMC article. - STAT1 and Its Crucial Role in the Control of Viral Infections.
Tolomeo M, Cavalli A, Cascio A. Tolomeo M, et al. Int J Mol Sci. 2022 Apr 7;23(8):4095. doi: 10.3390/ijms23084095. Int J Mol Sci. 2022. PMID: 35456913 Free PMC article. Review. - Human Mcm proteins at a replication origin during the G1 to S phase transition.
Schaarschmidt D, Ladenburger EM, Keller C, Knippers R. Schaarschmidt D, et al. Nucleic Acids Res. 2002 Oct 1;30(19):4176-85. doi: 10.1093/nar/gkf532. Nucleic Acids Res. 2002. PMID: 12364596 Free PMC article. - Role of STAT-1 and STAT-3 in ischaemia/reperfusion injury.
Stephanou A. Stephanou A. J Cell Mol Med. 2004 Oct-Dec;8(4):519-25. doi: 10.1111/j.1582-4934.2004.tb00476.x. J Cell Mol Med. 2004. PMID: 15601580 Free PMC article. Review.
References
- Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. - PubMed
- Leaman D W, Leung S, Li X, Stark G R. FASEB J. 1996;10:1578–1588. - PubMed
- Darnell J E., Jr Science. 1997;277:1630–1635. - PubMed
- Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. - PubMed
- O'Shea J J. Immunity. 1997;7:1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous