Selective formation of Sed5p-containing SNARE complexes is mediated by combinatorial binding interactions - PubMed (original) (raw)
Selective formation of Sed5p-containing SNARE complexes is mediated by combinatorial binding interactions
M M Tsui et al. Mol Biol Cell. 2001 Mar.
Free PMC article
Abstract
Sed5p is the only syntaxin family member required for protein transport through the yeast Golgi and it is known to bind up to nine other soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins in vivo. We describe in vitro binding experiments in which we identify ternary and quaternary Sed5p-containing SNARE complexes. The formation of SNARE complexes among these endoplasmic reticulum- and Golgi-localized proteins requires Sed5p and is syntaxin-selective. In addition, Sed5p-containing SNARE complexes form selectively and this selectivity is mediated by Sed5p-containing intermediates that discriminate among subsequent binding partners. Although many of these SNAREs have overlapping distributions in vivo, the SNAREs that form complexes with Sed5p in vitro reflect their functionally distinct locales. Although SNARE-SNARE interactions are promiscuous and a single SNARE protein is often found in more than one complex, both the biochemical as well as genetic analyses reported here suggest that this is not a result of nonselective direct substitution of one SNARE for another. Rather our data are consistent with the existence of multiple (perhaps parallel) trafficking pathways where Sed5p-containing SNARE complexes play overlapping and/or distinct functional roles.
Figures
Figure 1
(A) Amino acid sequence alignments of the heptad repeat/core domains of the SNAREs used in these studies. The a and d positions of the predicted hydrophobic layers (heptad repeats) are indicated above the arrows and the amino acids at these positions are highlighted in bold. The thick arrow indicates the position of the d amino acid, which has been used to define proteins as either R- or Q-SNAREs (Fasshauer et al., 1998). This position also corresponds to the zero ionic layer in the structure of the neuronal SNARE complex (Sutton et al., 1998). SNARE proteins are aligned with their putative family members: the syntaxins (Sed5p, Pep12p, Vam3p, and Tlg1p), the Bet1 family (Sft1p, Bet1p, and Vti1p), the Gos1p family (Gos1p and Bos1p), and the synaptobrevin family (Ykt6p, Sec22p, and Snc2p). The position of the amino acids in each of the proteins used in the alignments, is indicated in parentheses. The assignment of SNAREs to a particular family is based in part on amino acid sequence similarity as well as on the behavior of individual SNARE proteins in in vitro mixing assays (see text for details). (B) Pairwise percentage of amino acid identities among the aligned SNARE family proteins as indicated in A.
Figure 2
Formation of binary and ternary SNARE complexes with GST-Sft1p and GST-Sed5p. Soluble lysates from bacterial cells expressing recombinant GST- or (His)6-tagged SNARE proteins (lacking their membrane anchor sequences) were mixed together and protein complexes isolated by affinity chromatography with glutathione Sepharose beads. Proteins were resolved by SDS-PAGE and stained with Coomassie brilliant blue. (A) GST-Sft1p binds to [Sed5p + Sec22p] (lane 1) and to [Sed5p] (lane 2), but binding to [Sec22p] is not detectable (lane 3). (B) GST-Sed5p binds [Sec2p + Bos1p] (lane 3) and [Sec22p] albeit weakly (lane 2) but not to [Bos1p] (lane 1). The star symbol (★) in Figure 2A denotes GST derived from proteolysis of GST-Sft1p. The dots either side of lane 2 in A highlight the presence of Sed5p.
Figure 3
Sed5p selectively forms ternary and quaternary complexes in vitro with the SNAREs Sft1p, Bet1p, Gos1p, Bos1p, Ykt6p, and Sec22p. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and recombinant (His)6-tagged SNAREs detected by immunostaining with the anti-(His)6 antibody BMG-His-1. Ternary complexes: GST-Sft1p was mixed with various combinations of two different (His)6-tagged SNARE proteins as indicated (A). GST-Bet1p was mixed with various combinations of two different (His)6-tagged SNARE proteins as indicated (B). Quaternary complexes: Either GST-Bet1p or GST-Sft1p was mixed with (His)6-Sed5p and various combinations of two other (His)6-tagged SNARE proteins as indicated (C). Immunoblot analysis of aliquots of soluble lysates from bacteria expressing the (His)6-SNAREs used in this experiment (corresponding to ∼5% of input) (D). (E) Longer exposure of a subsection of B reveals some association of Ykt6p with [Bet1p + Sed5p] and of Bos1p with [Bet1p + Sed5p]. Note that in the absence of Sed5p no detectable binding is observed between either GST-Sft1p or GST-Bet1p and other SNAREs (A and B, lanes 5–8).
Figure 4
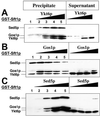
The formation of quaternary SNARE complexes occurs sequentially and only in the presence of Sed5p. In A–C, the relative amount of a single SNARE was sequentially increased (lanes 2–5), whereas the amount of the other three SNAREs remained constant. SNARE complex formation was monitored following affinity purification of GST-Sft1p on glutathione Sepharose beads (Precipitate) and immunostaining with an anti-(His)6 antibody. Aliquots of supernatants, removed following incubation of the mixtures with reduced glutathione beads (∼5% of total volume), were also analyzed by immunostaining (Supernatant). (A) Effect of increasing concentrations of Ykt6p. Note that (His)6-Yktp stimulates (weakly) the formation of the ternary complex of [Sed5p + Gos1p + Sft1p] (lane 2). (B) Gos1p. Progressively increasing (His)6-Gos1p results in the eventual reduction of the amount of GST-Sft1p quaternary complex (compare lanes 2–5). (C) Sed5p. Neither binary nor ternary Sft1p-SNARE complexes (with Gos1p or Ykt6p) form in the absence of Sed5p (lane 1).
Figure 5
Vti1p forms a quaternary complex with Sed5p, Tlg1p, and Ykt6p. Mixing assays and immunostaining were performed as described in MATERIALS AND METHODS. (A) GST-Vti1p was mixed with (His)6-Sed5p and various combinations of six other (His)6-tagged SNARE proteins as indicated. (B) GST-Sed5p was mixed with either (His)6-Ykt6p or (His)6-Tlg1p or both (His)6-tagged proteins as indicated. Note that Ykt6p facilitates the association of Tlg1p with Sed5p. (C) GST-Sft1p was mixed in various combinations with four different (His)6-tagged SNARE proteins as indicated. (D) Immunostaining of aliquots of soluble lysates from bacteria expressing the (His)6-SNAREs used in this experiment (∼5% of input). For the relative expression levels of (His)6-Vti1p and (His)6-Bos1p, see Figure 6B.
Figure 6
Snc2p forms a quaternary complex with Sed5p, Vti1p, and Tlg1p. Mixing assays and immunostaining were performed as described in the MATERIALS AND METHODS. (A) GST-Snc2p and (His)6-Sed5p were mixed with various combinations of five different (His)6-tagged SNARE proteins as indicated. (B) Immunostaining of aliquots of soluble lysates from bacteria expressing the (His)6-SNAREs used in this experiment (∼5% of input).
Figure 7
Vam3p and Pep12p do not form quaternary complexes with [Ykt6p + Gos1p + Sft1p], [Ykt6p + Tlg1p + Vti1p] or [Snc2p + Tlg1p + Vti1p]. Mixing assays and immunostaining were preformed as described in MATERIALS AND METHODS. (A) GST-Sft1p was mixed with either the syntaxin (His)6-Sed5p, (His)6-Vam3p, (His)6-Pep12p or (His)6-Tlg1p together with the (His)6-SNAREs Ykt6p and Gos1p. (B) GST-Vti1p was mixed with either (His)6-Sed5p, (His)6-Vam3p or (His)6-Pep12p along with the (His)6-SNAREs Tlg1p and Ykt6p. (C) GST-Snc2p was mixed with (His)6-Sed5p, (His)6-Vam3p, or (His)6-Pep12p together with (His)6-Vti1p and (His)6-Tlg1p. (D) GST-Vam7p was mixed with either (His)6-Vam3p or (His)6-Pep12p together with (His)6-Vti1p and (His)6-Tlg1p. (E) Immunostaining of aliquots of soluble lysates from bacteria expressing the (His)6-SNAREs used in this experiment (∼5% of input).
Figure 8
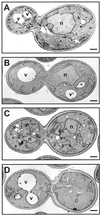
Electron micrographs of sft1Δ cells containing either the 2 μ SFT1 (SARY160) 2 μ_BET1_ (SARY181), or 2 μ SNC2 plasmids (SARY208). Yeast strains were grown at 25°C for 4 h and permanganate fixed to accentuate membranes. (A) sft1Δ 2 μ SFT1 (SARY160). Overexpression of Sft1p results in some vacuolar fragmentation as well as the accumulation of some dark membranous structures. (B) sft1Δ 2 μ_BET1_ (SARY181). Cells are indistinguishable from wild-type (D). (C) sft1Δ 2 μ SNC2 cells (SARY208). Note no significant differences in amount of ER membranes (compare with A, B, and D) but fragmentation of the vacuoles and the appearance of numerous dark membranous structures. (D) Wild-type parental strain (SEY6210, Table 1). The bar in the bottom right-hand side of each micrograph corresponds to 0.5 μm. N, nucleus; V, vacuole. The doubling times for each of the strains at 25°C were pSFT1 sft1Δ (SARY160), 2.0 h; pBET1_sft1Δ_ (SARY181), 2.3 h; pSNC2_sft1Δ_ (SARY 208), 6.7 h; and wild-type (SEY6210), 1.9 h (Table 1).
Figure 9
gos1Δ and sec22-3 do not display synthetic lethal interactions. Following sporulation of the heterozygous double mutant diploid (SARY327, Table 1), tetrads were dissected on YEPD plates containing 1 M sorbitol and incubated at 25°C for 5 d. (A) Plates shown in B–D were incubated at the indicated temperatures after replica plating (A) for 2 d. Tetrads 2, 4, and 6 are the parental ditype and tetrads 1, 3, 5, and 7 are tetratype. Spores 1a, 3b, 5c, and 7d correspond to the_gos1Δ_ sec22-3 double mutant progeny. The same result was also obtained for the gos1Δ and_sec22Δ_ mutations (data not shown).
Similar articles
- Yeast Golgi SNARE interactions are promiscuous.
Tsui MM, Banfield DK. Tsui MM, et al. J Cell Sci. 2000 Jan;113 ( Pt 1):145-52. doi: 10.1242/jcs.113.1.145. J Cell Sci. 2000. PMID: 10591633 - Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic.
Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG. Lupashin VV, et al. Mol Biol Cell. 1997 Dec;8(12):2659-76. doi: 10.1091/mbc.8.12.2659. Mol Biol Cell. 1997. PMID: 9398683 Free PMC article. - Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport.
Peng R, Gallwitz D. Peng R, et al. EMBO J. 2004 Oct 13;23(20):3939-49. doi: 10.1038/sj.emboj.7600410. Epub 2004 Sep 16. EMBO J. 2004. PMID: 15372079 Free PMC article. - SNAREs and membrane fusion in the Golgi apparatus.
Nichols BJ, Pelham HR. Nichols BJ, et al. Biochim Biophys Acta. 1998 Aug 14;1404(1-2):9-31. doi: 10.1016/s0167-4889(98)00044-5. Biochim Biophys Acta. 1998. PMID: 9714710 Review. - Stx5-Mediated ER-Golgi Transport in Mammals and Yeast.
Linders PT, Horst CV, Beest MT, van den Bogaart G. Linders PT, et al. Cells. 2019 Jul 26;8(8):780. doi: 10.3390/cells8080780. Cells. 2019. PMID: 31357511 Free PMC article. Review.
Cited by
- Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity.
Parlati F, Varlamov O, Paz K, McNew JA, Hurtado D, Söllner TH, Rothman JE. Parlati F, et al. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5424-9. doi: 10.1073/pnas.082100899. Proc Natl Acad Sci U S A. 2002. PMID: 11959998 Free PMC article. - Organization of SNAREs within the Golgi stack.
Malsam J, Söllner TH. Malsam J, et al. Cold Spring Harb Perspect Biol. 2011 Oct 1;3(10):a005249. doi: 10.1101/cshperspect.a005249. Cold Spring Harb Perspect Biol. 2011. PMID: 21768609 Free PMC article. Review. - Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes.
Peng R, Gallwitz D. Peng R, et al. J Cell Biol. 2002 May 13;157(4):645-55. doi: 10.1083/jcb.200202006. Epub 2002 May 6. J Cell Biol. 2002. PMID: 11994317 Free PMC article. - MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae.
Dou X, Wang Q, Qi Z, Song W, Wang W, Guo M, Zhang H, Zhang Z, Wang P, Zheng X. Dou X, et al. PLoS One. 2011 Jan 24;6(1):e16439. doi: 10.1371/journal.pone.0016439. PLoS One. 2011. PMID: 21283626 Free PMC article. - Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p.
Bracher A, Weissenhorn W. Bracher A, et al. EMBO J. 2002 Nov 15;21(22):6114-24. doi: 10.1093/emboj/cdf608. EMBO J. 2002. PMID: 12426383 Free PMC article.
References
- Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. - PubMed
- Banfield DK, Lewis MJ, Pelham HRB. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases