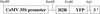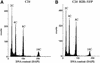Dynamic analyses of the expression of the HISTONE::YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains - PubMed (original) (raw)
Comparative Study
Dynamic analyses of the expression of the HISTONE::YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains
C Boisnard-Lorig et al. Plant Cell. 2001 Mar.
Abstract
During early seed development, nuclear divisions in the endosperm are not followed by cell division, leading to the development of a syncytium. The simple organization of the Arabidopsis endosperm provides a model in which to study the regulation of the cell cycle in relation to development. To monitor nuclear divisions, we constructed a HISTONE 2B::YELLOW FLUORESCENT PROTEIN gene fusion (H2B::YFP). To validate its use as a vital marker for chromatin in plants, H2B::YFP was expressed constitutively in Arabidopsis. This enabled the observation of mitoses in living root meristems. H2B::YFP was expressed specifically in Arabidopsis syncytial endosperm by using GAL4 transactivation. Monitoring mitotic activity in living syncytial endosperm showed that the syncytium was organized into three domains in which nuclei divide simultaneously with a specific time course. Each mitotic domain has a distinct spatiotemporal pattern of mitotic CYCLIN B1;1 accumulation. The polar spatial organization of the three mitotic domains suggests interactions between developmental mechanisms and the regulation of the cell cycle.
Figures
Figure 1.
H2B::YFP Fusion Constructs. Scheme of the T-DNA left border region of transfer vector pBI121 in which the EcoRI is deleted. Fusion was created by the insertion of the H2B cDNA between the BamHI and EcoRI sites of either pBI121 or pBIB. CaMV 35S promoter, 35S promoter from the cauliflower mosaic virus; H2B, histone 2B from Arabidopsis amplified by reverse transcription–polymerase chain reaction and homolog to ATH2B (GenBank accession number Y07745); YFP, modified sequence of yellow fluorescent protein (Haseloff, 1999); 3′, nopaline synthase terminator.
Figure 2.
Analysis of the Repartition of Cell Cycle Phases in Plants Expressing H2B::YFP. Nuclei of root cells were purified and labeled with DAPI, and fluorescence was analyzed by FACS. The phases of the cell cycle were identified as 2C (G1) or 4C (G2) of nuclear DNA content. Nuclei with higher DNA content (8C and 16C) have undergone endoreduplication. Endoreduplication and the repartition of cells between the phases of the cell cycle were not significantly different between plants that did not (A) or did (B) express the H2B::YFP fusion protein.
Figure 3.
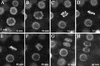
Colocalization of H2B::YFP Fluorescence and DNA Stained with Propidium Iodide in Fixed Root Meristems. (A) Confocal section of a root meristem. H2B::YFP fluorescence is shown in green. A more detailed image of one nucleus (inset) shows the nucleolus (n) and dense chromatin foci (arrowheads). (B) DNA stained with propidium iodide (PI) is shown in red. Condensed chromosomes during late anaphase are visible (arrow). PI fluorescence corresponding to organelle DNA is visible as spots around each nucleus. (C) The H2B::YFP signal colocalizes with the PI signal in nuclei during interphase and mitosis.
Figure 4.
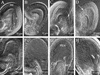
Dynamics of the Nuclear Structure and of the Chromatin from G2 Phase to G1 Phase in Living Root Epidermal Cells. (A) The cell at the bottom of the optical section was observed by using time lapse confocal microscopy during mitosis. The time elapsed from the first recognizable sign of nuclear envelope breakdown is indicated. Chromatin condensation foci are visible at the nuclear envelope and at the surface of the nucleolus. (B) In prophase, the nucleus loses its spherical shape and shows condensed chromatin aggregates. (C) These aggregates further condense into recognizable chromosomes. (D) Chromosomes align along the metaphase plate. (E) and (F) During anaphase, chromosomes separate (E) and migrate to opposite poles of the cell (F). (G) During telophase, chromosomes decondense. (H) A pair of cells results from the division of the G2 cell shown in (A).
Figure 5.
Main Developmental Stages of the Syncytial Endosperm. Whole mounts of cleared seed were observed by using differential interference contrast (DIC) optics. (A) The zygotic endosperm is tube shaped and surrounded by four to five cell layers of integuments (int). It is separated from the micropyle (mic) by the zygote (z) and the degenerating synergids (s). At the opposite pole, the chalazal proliferating tissue (chz) separates the endosperm from the vascular elements of the funiculus. (B) The first nuclear division is not followed by the division of the central cell, which becomes binucleate. At this stage, one nucleus is located closer to the zygote and the other is closer to the chalazal pole. Both nuclei occupy a central position in the endosperm tube. (C) The four-nuclei–stage endosperm is the result of the second round of syncytial nuclei division. One nucleus migrates to the vicinity of the zygote (arrow). (D) The third round of nuclear division takes place perpendicular to the polar axis of the now enlarged endosperm tube. A pair of nuclei is located close to the zygote, and one or two nuclei are located at the chalazal pole (arrow). (E) At the two-cell embryo stage, one to four nuclei occupy the cleft formed above the chalaza. Those nuclei are usually larger than the nuclei located in the periphery and are characterized by large nucleoli. (F) Quadrant-stage embryo with stage VIII endosperm. (G) The globular embryo protrudes inside the endosperm cavity and is surrounded by a layer of endosperm nuclei. Three morphologically distinct zones can be identified: the micropylar pole occupied by a dense cytoplasmic embryo-surrounding region (MCE); the peripheral endosperm (PEN), which is the largest domain in terms of total number of nuclei and total surface area; and, at the chalazal pole, a pocket that contains few nuclei and constitutes the chalazal endosperm (CZE). The PEN is composed of one layer of nuclear cytoplasmic domains evenly spaced along the entire surface of the endosperm. (H) The endosperm becomes cellularized at the embryo torpedo stage. A section of the surface of the endosperm shows hexagonal and pentagonal endosperm cell sections.
Figure 6.
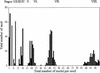
Developmental Stages of the Syncytial Endosperm as Defined by the Total Number of Nuclei. A large population of developing seed was divided into classes defined by the total number of endosperm nuclei. It appears that seed are divided into subpopulations that correspond to distinct stages of development.
Figure 7.
Transactivation of H2B::YFP in the Endosperm. (A) The GAL4::VP16 enhancer trap line KS22 expresses the ER-targeted variant mGFP5 in the endosperm in the plant seed. This outlines the ER-rich MCE and CZE and the nuclear cytoplasmic domain in the PEN. Nuclei appear as black discs surrounded by brighter nuclear envelopes. (B) The KS22 line was transformed with promUAS-H2B::YFP. This causes specific labeling of chromatin in endosperm nuclei. All nuclei appear similar in diameter and intensity of labeling, with the exception of larger and brighter nuclei in the CZE.
Figure 8.
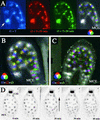
Time Lapse Observation of Nuclear Division in Mitotic Domains of the Syncytial Endosperm. Time lapse imaging of nuclei in KS22 seed expressing H2B::YFP enabled us to monitor nuclear division during endosperm development. Optical sections were recorded every 10 min for 12 hr. Only three consecutive sections are displayed, corresponding to the time at which nuclear divisions were observed. Each section is color coded with the fundamental colors red, blue, and green. The superposition of the three images clearly shows nuclei that do not divide (white) and nuclei that undergo mitosis (color). (A) Transition from stage V to stage VI. Two large nuclei in the CZE do not participate in the synchronous division that affects all other nuclei in the endosperm (arrow). (B) Transition from stage VI to stage VII. Superposition of three color-coded consecutive sections obtained every 10 min during a 12-hr time lapse recording. CZE nuclei do not divide and appear white. White nuclei in the PEN divide perpendicular to the plane of observation. PEN synchronous nuclear division precedes by 10 min MCE nuclear division, resulting in different color sequences. (C) A similar recording of nuclear division in a stage VII endosperm shows that MCE nuclear division takes place independently from coordinated PEN mitoses. MCE nuclei thus appear white, as do CZE nuclei. A few PEN nuclei located close to the chalazal pole do not divide until the next period (data not shown). This results from the fact that nuclear division takes place in the PEN as a wave from the MCE toward the CZE. (D) A series of consecutive optical sections from a 14-hr time lapse recording of the transition from stage VII to stage VIII. Negative images of optical sections are displayed. The endosperm is observed from the chalazal pole. CZE nuclei are larger and brighter than all other nuclei and do not experience mitosis when it is observed in the PEN. A wave of nuclear division crosses the PEN, as indicated by the arrow. The four nuclei closest to the CZE large nuclei do not divide and instead become part of the CZE.
Figure 9.
Mitotic Domains in Arabidopsis Endosperm Observed in Fixed Seed. Whole mounts of cleared seed are observed with DIC optics. (A) Endosperm at the transition between stages VI and VII with a two-cell embryo. Metaphase plates are detected in all nuclei with the exception of the two large nuclei of the CZE. (B) Transition from stage VIII to stage IX. All nuclei in the PEN are in anaphase, whereas nuclei in the MCE are in interphase. (C) Endosperm at stage VIII with a dermatogen embryo. Nuclei in the MCE are in metaphase, whereas nuclei in the PEN are in interphase.
Figure 10.
CYCLIN B1;1 Is Differentially Expressed in the Syncytial Endosperm Mitotic Domains MCE, PEN, and CZE. Whole mounts of cleared FA4C seed are observed after GUS staining. This reflects the expression of the CyclinB1;1::GUS fusion protein. (A) Endosperm at stage VI with a two-cell embryo. GUS stain is present in all nuclei of the PEN but not in the CZE occupied by four nuclei (only three nuclei are visible). (B) A stage VIII endosperm with GUS expression restricted to the PEN. The octant embryo shows a strong GUS signal on two cells of the embryo proper. (C) Endosperm at stage VII. The GUS stain is localized in the MCE of a two-cell embryo.
Comment in
- A brief tour of the cell cycle.
Eckardt NA. Eckardt NA. Plant Cell. 2001 Mar;13(3):449-51. doi: 10.1105/tpc.13.3.449. Plant Cell. 2001. PMID: 11251087 Free PMC article. No abstract available.
Similar articles
- Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation.
Day RC, Herridge RP, Ambrose BA, Macknight RC. Day RC, et al. Plant Physiol. 2008 Dec;148(4):1964-84. doi: 10.1104/pp.108.128108. Epub 2008 Oct 15. Plant Physiol. 2008. PMID: 18923020 Free PMC article. - ANAPHASE PROMOTING COMPLEX/CYCLOSOME-mediated cyclin B1 degradation is critical for cell cycle synchronization in syncytial endosperms.
Guo L, Jiang L, Lu XL, Liu CM. Guo L, et al. J Integr Plant Biol. 2018 Jun;60(6):448-454. doi: 10.1111/jipb.12641. Epub 2018 May 8. J Integr Plant Biol. 2018. PMID: 29424475 - The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation.
Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH. Yu Y, et al. Plant Cell. 2003 Dec;15(12):2763-77. doi: 10.1105/tpc.015990. Epub 2003 Nov 13. Plant Cell. 2003. PMID: 14615597 Free PMC article. - Endosperm development.
Berger F. Berger F. Curr Opin Plant Biol. 1999 Feb;2(1):28-32. doi: 10.1016/s1369-5266(99)80006-5. Curr Opin Plant Biol. 1999. PMID: 10047564 Review. - Fungal syncytia.
Jalihal AP, Gladfelter AS. Jalihal AP, et al. Curr Biol. 2025 Jun 9;35(11):R490-R495. doi: 10.1016/j.cub.2025.01.054. Curr Biol. 2025. PMID: 40494302 Review.
Cited by
- New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton.
Wang L, Ruan YL. Wang L, et al. Plant Physiol. 2012 Oct;160(2):777-87. doi: 10.1104/pp.112.203893. Epub 2012 Aug 3. Plant Physiol. 2012. PMID: 22864582 Free PMC article. - Tools for Assessing Cell-Cycle Progression in Plants.
Echevarría C, Gutierrez C, Desvoyes B. Echevarría C, et al. Plant Cell Physiol. 2021 Nov 10;62(8):1231-1238. doi: 10.1093/pcp/pcab066. Plant Cell Physiol. 2021. PMID: 34021583 Free PMC article. Review. - High frequency, cell type-specific visualization of fluorescent-tagged genomic sites in interphase and mitotic cells of living Arabidopsis plants.
Matzke AJ, Watanabe K, van der Winden J, Naumann U, Matzke M. Matzke AJ, et al. Plant Methods. 2010 Jan 19;6:2. doi: 10.1186/1746-4811-6-2. Plant Methods. 2010. PMID: 20148117 Free PMC article. - Eukaryotic cells and their cell bodies: Cell Theory revised.
Baluska F, Volkmann D, Barlow PW. Baluska F, et al. Ann Bot. 2004 Jul;94(1):9-32. doi: 10.1093/aob/mch109. Epub 2004 May 20. Ann Bot. 2004. PMID: 15155376 Free PMC article. Review. - BABY BOOM regulates early embryo and endosperm development.
Chen B, Maas L, Figueiredo D, Zhong Y, Reis R, Li M, Horstman A, Riksen T, Weemen M, Liu H, Siemons C, Chen S, Angenent GC, Boutilier K. Chen B, et al. Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2201761119. doi: 10.1073/pnas.2201761119. Epub 2022 Jun 16. Proc Natl Acad Sci U S A. 2022. PMID: 35709319 Free PMC article.
References
- Bajer, A.S. (1958). Cine-micrographic studies on mitosis in endosperm. IV. Exp. Cell Res. 14, 245–256. - PubMed
- Bajer, A.S., and Molè-Bajer, J. (1972). Spindle dynamics and chromosome movements. Int. Rev. Cytol. 3 (suppl.), 34.–65.
- Bechtold, C., Ellis, P., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris Life Sci. 316, 1194–1199.
- Berger, F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2, 28–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources