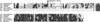Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus - PubMed (original) (raw)
doi: 10.1105/tpc.13.3.645.
R G Olmstead, K L Adams, J D Palmer, N T Lao, L Heggie, T A Kavanagh, J M Hibberd, J C Gray, C W Morden, P J Calie, L S Jermiin, K H Wolfe
Affiliations
- PMID: 11251102
- PMCID: PMC135507
- DOI: 10.1105/tpc.13.3.645
Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus
R S Millen et al. Plant Cell. 2001 Mar.
Abstract
We used DNA sequencing and gel blot surveys to assess the integrity of the chloroplast gene infA, which codes for translation initiation factor 1, in >300 diverse angiosperms. Whereas most angiosperms appear to contain an intact chloroplast infA gene, the gene has repeatedly become defunct in approximately 24 separate lineages of angiosperms, including almost all rosid species. In four species in which chloroplast infA is defunct, transferred and expressed copies of the gene were found in the nucleus, complete with putative chloroplast transit peptide sequences. The transit peptide sequences of the nuclear infA genes from soybean and Arabidopsis were shown to be functional by their ability to target green fluorescent protein to chloroplasts in vivo. Phylogenetic analysis of infA sequences and assessment of transit peptide homology indicate that the four nuclear infA genes are probably derived from four independent gene transfers from chloroplast to nuclear DNA during angiosperm evolution. Considering this and the many separate losses of infA from chloroplast DNA, the gene has probably been transferred many more times, making infA by far the most mobile chloroplast gene known in plants.
Figures
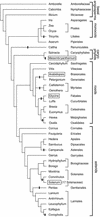
Figure 1.
Losses of Chloroplast infA during Angiosperm Evolution Based on Analysis of Sequenced infA Loci. Shown are all angiosperms whose chloroplast infA loci have been sequenced, except that only one representative of the 17 sequenced Solanaceae species is shown. Black bars indicate lineages with chloroplast infA pseudogenes or complete loss. The branching order and taxonomic classifications are based on Soltis et al. (1999) and Olmstead et al. (2000). Boxed names indicate species from which a nuclear infA gene was identified.
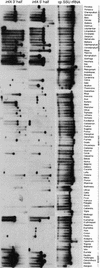
Figure 2.
Evidence for Loss of Chloroplast infA from DNA Gel Blot Hybridization. The three probes named at top were hybridized sequentially to a filter containing BamHI digests of total DNA from 280 angiosperms, 96 of which are shown here. Note that some infA genes have a BamHI site near their middle. SSU, small subunit.
Figure 3.
Losses of Chloroplast infA during Angiosperm Evolution Based on DNA Gel Blot Hybridization Results. The infA probes used were both from Antirrhinum. Plants with significantly diminished or no hybridization are shaded (see Figure 2 and text). Plant names beside the tree alternate in two columns. Loss lineages are shown in boldface on the tree and are marked with bullets. Bull's-eye bullets indicate the eight loss lineages for which loss was confirmed by sequencing of chloroplast infA from at least one member of the lineage. Within rosids, the sequenced member(s) was included on the blots; for the other four sequence-validated loss lineages, a different member of the loss lineage was sequenced than was on the blots. In particular, Tricyrtis (Figure 2) is the sister taxon to Smilax relative to all other taxa on the blots, Caltha is likewise sister to the _Clematis_-Ranunculus clade, Pentas is sister to Galium, and Campanula belongs within the _Lobelia_-Trachelium clade.
Figure 4.
Multiple Alignment of IF1 Protein Sequences from the Four Nuclear infA Genes and Two Representative Chloroplast Genes. The alignment was made using ClustalW 1.8 (Thompson et al., 1994), with the most common residue at each position highlighted. Predicted chloroplast transit peptides are shown by lowercase letters, and their predicted cleavage sites (Emanuelsson et al., 1999) are marked by large X symbols. The alignment of the transit peptides is not meant to indicate that they share a common ancestor (see text). cp, chloroplast; nuc, nuclear.
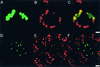
Figure 5.
Localization of Fusion Proteins Encoded by Nuclear InfA_-gfp Constructs to Chloroplasts. The soybean InfA-gfp and Arabidopsis InfA-gfp were introduced into epidermal cells of tobacco and Arabidopsis using microprojectile bombardment. The localization of GFP in individual transformed cells was detected by confocal laser scanning microscopy 3 days after bombardment. (A) to (C) p_GmInfA-gfp in a tobacco guard cell. The images show two guard cells; the upper cell shows expression of p_GmInfA-gfp_. GFP fluorescence (A). Chlorophyll fluorescence (B). Merged images (C). Bar in (C) = 5 μm. (D) to (F) Arabidopsis epidermal pavement cells with one cell expressing p_AtInfA-gfp._ The chloroplasts in epidermal pavement cells are smaller than those in guard cells. GFP fluorescence (D). Chlorophyll fluorescence (E). Merged images (F). Bar in (F) = 10 μm.
Figure 6.
Phylogenetic Analysis of Nuclear and Chloroplast infA Sequences. The diagram summarizes the maximum likelihood trees found when each of the four nuclear infA sequences was added individually to a constrained topology (the species phylogeny from Figure 1) for the 21 chloroplast sequences. The arrows indicate the most likely placement of the tomato (Solanum), soybean (Glycine), ice plant (Mesembryanthemum), and Arabidopsis nuclear sequences, and all branch lengths are drawn to scale horizontally. Mesembryanthemum cp denotes the ice plant chloroplast pseudogene. The branch lengths for the chloroplast sequences were identical in the four trees, allowing them to be merged. cp, chloroplast; nuc, nuclear.
Similar articles
- Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts.
Adams KL, Daley DO, Whelan J, Palmer JD. Adams KL, et al. Plant Cell. 2002 Apr;14(4):931-43. doi: 10.1105/tpc.010483. Plant Cell. 2002. PMID: 11971146 Free PMC article. - The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple': organization and phylogenetic relationships to other angiosperms.
Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. Bausher MG, et al. BMC Plant Biol. 2006 Sep 30;6:21. doi: 10.1186/1471-2229-6-21. BMC Plant Biol. 2006. PMID: 17010212 Free PMC article. - Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus.
Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H. Jansen RK, et al. Mol Biol Evol. 2011 Jan;28(1):835-47. doi: 10.1093/molbev/msq261. Epub 2010 Oct 8. Mol Biol Evol. 2011. PMID: 20935065 Free PMC article. - Gene transfer from organelles to the nucleus: frequent and in big chunks.
Martin W. Martin W. Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8612-4. doi: 10.1073/pnas.1633606100. Epub 2003 Jul 14. Proc Natl Acad Sci U S A. 2003. PMID: 12861078 Free PMC article. Review. No abstract available. - Nuclear DNA amounts in angiosperms: progress, problems and prospects.
Bennett MD, Leitch IJ. Bennett MD, et al. Ann Bot. 2005 Jan;95(1):45-90. doi: 10.1093/aob/mci003. Ann Bot. 2005. PMID: 15596457 Free PMC article. Review.
Cited by
- Endosybiotic evolution in action: Real-time observations of chloroplast to nucleus gene transfer.
Lloyd AH, Timmis JN. Lloyd AH, et al. Mob Genet Elements. 2011 Sep;1(3):216-220. doi: 10.4161/mge.1.3.17947. Epub 2011 Sep 1. Mob Genet Elements. 2011. PMID: 22479690 Free PMC article. - Comparative and phylogenetic analyses of nine complete chloroplast genomes of Orchidaceae.
Liu L, Du J, Liu Z, Zuo W, Wang Z, Li J, Zeng Y. Liu L, et al. Sci Rep. 2023 Dec 4;13(1):21403. doi: 10.1038/s41598-023-48043-2. Sci Rep. 2023. PMID: 38049440 Free PMC article. - Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development.
Motohashi R, Yamazaki T, Myouga F, Ito T, Ito K, Satou M, Kobayashi M, Nagata N, Yoshida S, Nagashima A, Tanaka K, Takahashi S, Shinozaki K. Motohashi R, et al. Plant Mol Biol. 2007 Jul;64(5):481-97. doi: 10.1007/s11103-007-9166-7. Epub 2007 Apr 21. Plant Mol Biol. 2007. PMID: 17450416 - Revisiting chloroplast genomic landscape and annotation towards comparative chloroplast genomes of Rhamnaceae.
Wanichthanarak K, Nookaew I, Pasookhush P, Wongsurawat T, Jenjaroenpun P, Leeratsuwan N, Wattanachaisaereekul S, Visessanguan W, Sirivatanauksorn Y, Nuntasaen N, Kuhakarn C, Reutrakul V, Ajawatanawong P, Khoomrung S. Wanichthanarak K, et al. BMC Plant Biol. 2023 Jan 28;23(1):59. doi: 10.1186/s12870-023-04074-5. BMC Plant Biol. 2023. PMID: 36707785 Free PMC article. - Chloroplast phylogenomic analysis provides insights into the evolution of the largest eukaryotic genome holder, Paris japonica (Melanthiaceae).
Yang L, Yang Z, Liu C, He Z, Zhang Z, Yang J, Liu H, Yang J, Ji Y. Yang L, et al. BMC Plant Biol. 2019 Jul 4;19(1):293. doi: 10.1186/s12870-019-1879-7. BMC Plant Biol. 2019. PMID: 31272375 Free PMC article.
References
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. - PubMed
- Allen, J.F. (1993). Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631. - PubMed
- Andrews, T.D., Jermiin, L.S., and Easteal, S. (1998). Accelerated evolution of cytochrome b in simian primates: Adaptive evolution in concert with other mitochondrial proteins? J. Mol. Evol. 47, 249–257. - PubMed
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases