Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest - PubMed (original) (raw)
Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest
A Constantinou et al. EMBO Rep. 2000 Jul.
Abstract
Individuals affected by the autosomal recessive disorder Werner's syndrome (WS) develop many of the symptoms characteristic of premature ageing. Primary fibroblasts cultured from WS patients exhibit karyotypic abnormalities and a reduced replicative life span. The WRN gene encodes a 3'-5' DNA helicase, and is a member of the RecQ family, which also includes the product of the Bloom's syndrome gene (BLM). In this work, we show that WRN promotes the ATP-dependent translocation of Holliday junctions, an activity that is also exhibited by BLM. In cells arrested in S-phase with hydroxyurea, WRN localizes to discrete nuclear foci that coincide with those formed by the single-stranded DNA binding protein replication protein A. These results are consistent with a model in which WRN prevents aberrant recombination events at sites of stalled replication forks by dissociating recombination intermediates.
Figures
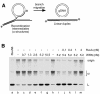
Fig. 1. Dissociation of recombination intermediates by WRN. (A) Schematic diagram indicating recombination intermediates (α-structures) and the expected products of branch migration. A region of heterology (1670 bp) at the distal end of the linear duplex blocks strand exchange and is represented by a filled box. The positions of the 32P-labels are indicated with asterisks. (B) Activity of WRN protein on recombination intermediates. Purified recombination intermediates (1.5 µM, expressed as a concentration of nucleotide residues) were incubated with: lane a, 200 nM RuvA and 100 nM RuvB; lanes c–g, 0.7–10.5 nM WRN; lanes i–m, 5.2 nM WRN and RuvA as indicated. In the reactions indicated, purified recombination intermediates were pre-incubated for 3 min on ice with RuvA before addition of WRN. The positions of the recombination intermediates (α) and linear duplex DNA (L) are indicated.
Fig. 2. Dependence on ATP hydrolysis. Dissociation reactions were carried out as described in the legend to Figure 1 using 1.5 µM 32P-labelled recombination intermediates and 5 nM WRN, in (A) standard buffer or (B) standard buffer in which ATP was replaced with 2 mM AMP-PNP. At the times indicated, aliquots were withdrawn and analysed by gel electrophoresis. (C) Quantification of reactions carried out in standard buffer containing ATP or AMP-PNP (A and B), or in the absence of NTPs (primary data not shown), as indicated. The amount of 32P-labelled linear duplex branch migration product was quantified by phosphoimaging and expressed as a percentage of the total radiolabel.
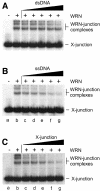
Fig. 3. Specificity of binding of WRN to synthetic Holliday junctions. Band-shift assays contained 7.3 nM WRN protein (indicated by +), 1 nM 32P-labelled synthetic Holliday junction (X-junction) and unlabelled DNA competitor as indicated. The competitors were (A) dsDNA, (B) ssDNA and (C) synthetic X-junction DNA, and were used at concentrations of 1.5 nM (lanes c), 3.1 nM (lanes d), 6.25 nM (lanes e), 12.5 nM (lanes f) and 25 nM (lanes g). DNA concentrations are expressed in moles of DNA substrate.
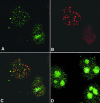
Fig. 4. Co-localization of WRN and RPA proteins in cells arrested in S-phase with HU. (A–C) Cells were arrested in S-phase by HU treatment as described. Following lysis, fixation and permeabilization, cells were incubated with anti-WRN (A; green) or anti-RPA antibodies (B; red). In >50% of the cells, WRN localizes in nuclear foci that coincide with the RPA foci, as illustrated in the superimposed images (C). The yellow colour results from the overlapping of the red and green foci. In cells incubated for the same length of time in the absence of HU, WRN shows the characteristic diffuse nucleolar (n) localization (D).
Similar articles
- Localization of the Bloom syndrome helicase to punctate nuclear structures and the nuclear matrix and regulation during the cell cycle: comparison with the Werner's syndrome helicase.
Gharibyan V, Youssoufian H. Gharibyan V, et al. Mol Carcinog. 1999 Dec;26(4):261-73. doi: 10.1002/(sici)1098-2744(199912)26:4<261::aid-mc5>3.0.co;2-a. Mol Carcinog. 1999. PMID: 10569803 - Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins.
Machwe A, Lozada EM, Xiao L, Orren DK. Machwe A, et al. BMC Mol Biol. 2006 Jan 13;7:1. doi: 10.1186/1471-2199-7-1. BMC Mol Biol. 2006. PMID: 16412221 Free PMC article. - Roles of the Bloom's syndrome helicase in the maintenance of genome stability.
Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID. Cheok CF, et al. Biochem Soc Trans. 2005 Dec;33(Pt 6):1456-9. doi: 10.1042/BST0331456. Biochem Soc Trans. 2005. PMID: 16246145 Review. - Asymmetry of DNA replication fork progression in Werner's syndrome.
Rodríguez-López AM, Jackson DA, Iborra F, Cox LS. Rodríguez-López AM, et al. Aging Cell. 2002 Oct;1(1):30-9. doi: 10.1046/j.1474-9728.2002.00002.x. Aging Cell. 2002. PMID: 12882351
Cited by
- The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae.
Wagner M, Price G, Rothstein R. Wagner M, et al. Genetics. 2006 Oct;174(2):555-73. doi: 10.1534/genetics.104.036905. Epub 2006 Jul 2. Genetics. 2006. PMID: 16816432 Free PMC article. - RecQ helicases: multiple structures for multiple functions?
Vindigni A, Hickson ID. Vindigni A, et al. HFSP J. 2009 Jun;3(3):153-64. doi: 10.2976/1.3079540. Epub 2009 Mar 18. HFSP J. 2009. PMID: 19949442 Free PMC article. - A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair.
Shor E, Weinstein J, Rothstein R. Shor E, et al. Genetics. 2005 Mar;169(3):1275-89. doi: 10.1534/genetics.104.036764. Epub 2005 Jan 16. Genetics. 2005. PMID: 15654096 Free PMC article. - Pathways of mammalian replication fork restart.
Petermann E, Helleday T. Petermann E, et al. Nat Rev Mol Cell Biol. 2010 Oct;11(10):683-7. doi: 10.1038/nrm2974. Epub 2010 Sep 15. Nat Rev Mol Cell Biol. 2010. PMID: 20842177 Review. - Probing the structure and function of polymerase θ helicase-like domain.
Vanson S, Li Y, Wood RD, Doublié S. Vanson S, et al. DNA Repair (Amst). 2022 Aug;116:103358. doi: 10.1016/j.dnarep.2022.103358. Epub 2022 Jun 14. DNA Repair (Amst). 2022. PMID: 35753097 Free PMC article. Review.
References
- Bennett R.J., Keck, J.L. and Wang, J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol., 289, 235–248. - PubMed
- Brosh R.M., Orren, D.K., Nehlin, J.O., Ravn, P.H., Kenny, M.K., Machwe, A. and Bohr, V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. - PubMed
- Chakraverty R.K. and Hickson, I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. BioEssays, 21, 286–294. - PubMed
- Eggleston A.K., Mitchell, A.H. and West, S.C. (1997) In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell, 89, 607–617. - PubMed
- Epstein C.J., Martin, G.M., Schultz, A.L. and Motulsky, A.G. (1966) Werner’s syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine, 45, 177–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources