Finding nuclear localization signals - PubMed (original) (raw)
Finding nuclear localization signals
M Cokol et al. EMBO Rep. 2000 Nov.
Abstract
A variety of nuclear localization signals (NLSs) are experimentally known although only one motif was available for database searches through PROSITE. We initially collected a set of 91 experimentally verified NLSs from the literature. Through iterated 'in silico mutagenesis' we then extended the set to 214 potential NLSs. This final set matched in 43% of all known nuclear proteins and in no known non-nuclear protein. We estimated that >17% of all eukaryotic proteins may be imported into the nucleus. Finally, we found an overlap between the NLS and DNA-binding region for 90% of the proteins for which both the NLS and DNA-binding regions were known. Thus, evolution seemed to have used part of the existing DNA-binding mechanism when compartmentalizing DNA-binding proteins into the nucleus. However, only 56 of our 214 NLS motifs overlapped with DNA-binding regions. These 56 NLSs enabled a de novo prediction of partial DNA-binding regions for approximately 800 proteins in human, fly, worm and yeast.
Figures
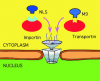
Fig. 1. Simplified scheme for nuclear import. Upon synthesis of nuclear proteins in the cytoplasm, e.g. the family of importins or transportins bind to the NLS. The complex importin/NLS protein (or transportin/protein) is then actively transported into the nucleus through nuclear pores involving the Ran GTPase cycle. Currently, this is the only known mechanism for nuclear import (Mattaj and Englmeier, 1998; Weis, 1998).
Fig. 2. NLS motif also used for DNA binding. Zoom into the interface between DNA and P55-C-fos proto-oncogene protein [note, the other parts of the amazing crystal structure of the complex with PDB code 1a02 (Chen et al., 1998) are not shown]. The coloured region corresponds to the residues RRERNKMAAAKSRNRRR. In fact, this motif is also contained in our data set of potential NLS motifs. Colouring scheme: basic residues shown in red, others in yellow. Graph created with RASMOL (Sayle and Milner-White, 1995).
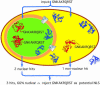
Fig. 3. Scheme for the concept of ‘in silico mutagenesis’. We started the search with the hypothetical motif GNKAKRQRST. We searched the data sets of proteins known to be nuclear and proteins known to be non-nuclear for the presence of this motif. In this particular example, two nuclear and one non-nuclear protein matched. Requiring 100% accuracy for all motifs, we did not include GNKAKRQRST into our data set of potential motifs. Note, the particular example was one of many failed attempts to generalize an experimental NLS.
Similar articles
- NLSdb: database of nuclear localization signals.
Nair R, Carter P, Rost B. Nair R, et al. Nucleic Acids Res. 2003 Jan 1;31(1):397-9. doi: 10.1093/nar/gkg001. Nucleic Acids Res. 2003. PMID: 12520032 Free PMC article. - NLSdb-major update for database of nuclear localization signals and nuclear export signals.
Bernhofer M, Goldberg T, Wolf S, Ahmed M, Zaugg J, Boden M, Rost B. Bernhofer M, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D503-D508. doi: 10.1093/nar/gkx1021. Nucleic Acids Res. 2018. PMID: 29106588 Free PMC article. - Nuclear Respiratory Factor 2β (NRF-2β) recruits NRF-2α to the nucleus by binding to importin-α:β via an unusual monopartite-type nuclear localization signal.
Hayashi R, Takeuchi N, Ueda T. Hayashi R, et al. J Mol Biol. 2013 Sep 23;425(18):3536-48. doi: 10.1016/j.jmb.2013.07.007. Epub 2013 Jul 13. J Mol Biol. 2013. PMID: 23856623 - Identification of nuclear import and export signals within Fli-1: roles of the nuclear import signals in Fli-1-dependent activation of megakaryocyte-specific promoters.
Hu W, Philips AS, Kwok JC, Eisbacher M, Chong BH. Hu W, et al. Mol Cell Biol. 2005 Apr;25(8):3087-108. doi: 10.1128/MCB.25.8.3087-3108.2005. Mol Cell Biol. 2005. PMID: 15798196 Free PMC article. - Nuclear localization signals: a driving force for nuclear transport of plasmid DNA in zebrafish.
Collas P, Aleström P. Collas P, et al. Biochem Cell Biol. 1997;75(5):633-40. Biochem Cell Biol. 1997. PMID: 9551185 Review.
Cited by
- CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis.
Huang XY, Niu J, Sun MX, Zhu J, Gao JF, Yang J, Zhou Q, Yang ZN. Huang XY, et al. Plant Cell. 2013 Feb;25(2):637-48. doi: 10.1105/tpc.112.107896. Epub 2013 Feb 12. Plant Cell. 2013. PMID: 23404887 Free PMC article. - Nuclear Effectors in Plant Pathogenic Fungi.
De Mandal S, Jeon J. De Mandal S, et al. Mycobiology. 2022 Sep 20;50(5):259-268. doi: 10.1080/12298093.2022.2118928. eCollection 2022. Mycobiology. 2022. PMID: 36404902 Free PMC article. Review. - Genome-scale metabolic model of the diatom Thalassiosira pseudonana highlights the importance of nitrogen and sulfur metabolism in redox balance.
van Tol HM, Armbrust EV. van Tol HM, et al. PLoS One. 2021 Mar 24;16(3):e0241960. doi: 10.1371/journal.pone.0241960. eCollection 2021. PLoS One. 2021. PMID: 33760840 Free PMC article. - HDAC4/5-HMGB1 signalling mediated by NADPH oxidase activity contributes to cerebral ischaemia/reperfusion injury.
He M, Zhang B, Wei X, Wang Z, Fan B, Du P, Zhang Y, Jian W, Chen L, Wang L, Fang H, Li X, Wang PA, Yi F. He M, et al. J Cell Mol Med. 2013 Apr;17(4):531-42. doi: 10.1111/jcmm.12040. Epub 2013 Mar 11. J Cell Mol Med. 2013. PMID: 23480850 Free PMC article. - Characterization and prediction of protein nucleolar localization sequences.
Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. Scott MS, et al. Nucleic Acids Res. 2010 Nov;38(21):7388-99. doi: 10.1093/nar/gkq653. Epub 2010 Jul 26. Nucleic Acids Res. 2010. PMID: 20663773 Free PMC article.
References
- Boulikas T. (1993) Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr., 3, 193–227. - PubMed
- Boulikas T. (1994) Putative nuclear localization signals (NLS) in protein transcription factors. J. Cell. Biochem., 55, 32–58. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases