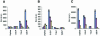Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300 - PubMed (original) (raw)
Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300
D Dornan et al. EMBO Rep. 2001 Feb.
Abstract
The N-terminal BOX-I domain of p53 containing a docking site for the negative regulator MDM2 and the positive effector p300, harbours two recently identified phosphorylation sites at Thr18 or Ser20O whose affect on p300 is undefined. Biochemical assays demonstrate that although MDM2 binding is inhibited by these phosphorylations, p300 binding is strikingly stabilized by Thr18 or Ser20 phosphorylation. Introducing EGFP-BOX-I domain peptides with an aspartate substitution at Thr18 or Ser20 induced a significant inhibition of endogenous p53-dependent transcription in cycling cells, in irradiated cells, as well as in cells transiently co-transfected with p300 and p53. In contrast an EGFP-wild-type BOX-I domain peptide stimulated p53 activity via inhibition of MDM2 protein binding. These results suggest that phosphorylation of p53 at Thr18 or Ser20 can activate p53 by stabilizing the p300-p53 complex and also identify a class of small molecular weight ligands capable of selective discrimination between MDM2- and p300-dependent activities.
Figures
Fig. 1. Phosphorylation stabilizes p300-p53 BOX-I peptide complexes. The binding of: (A) full-length p300 protein, (B) truncated p300 (1135–2414) and (C) full-length MDM2 protein, to the indicated biotinylated-peptides was analysed by ELISA as described in the Methods. The amount of p300 or MDM2 protein bound are represented as relative light units (RLUs). The amount of biotinylated peptide titrated onto streptavidin-coated ELISA surfaces is indicated in this Figure, and the other Figures where ELISA is used, as follows: dark blue bars, 1 ng; brown bars, 0.1 ng; yellow bars, 0.01 ng and light blue bars, 0 ng.
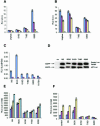
Fig. 2. In vivo inhibition of endogenous p53-dependent transcription by phospho-peptide mimetics. (A) p300 and (B) MDM2 binding in vitro to aspartate-substituted BOX-I domain peptides. The binding of p300 protein and MDM2 protein, to biotinylated-peptides substituted with aspartate at the indicated positions was as described in Figure 1. (C). EGFP-Asp18 and Asp20 peptide fusion proteins inhibit p53-dependent transactivation in vivo. EGFP-constructs or the mutant p53HIS175 allele (100 ng) were transiently transfected with 2 µg p21-Luc or 2 µg control-Luc and 1 µg control-β-Gal-reporter into cycling A375 cells, and the cells harvested 24 h post-transfection. p53-dependent activity (RLUs) is expressed as a ratio of p21-luciferase activity (dark blue bars) or control-luciferase activity (brown bars) to the internal transfection control (β-Gal). (D). Expression levels of EGFP-peptide fusion proteins in A375 cells. Lysates from cells transfected with the indicated EGFP-peptide fusion constructs, as described in Figure 2C, were immunoblotted with antibodies to EGFP to determine the relative level of each fusion protein expressed in cells transfected with the 2 µg p21-Luc or control-Luc vectors. (E) p300 can recover p53 activity in cells co-transfected with the inhibitory EGFP-Asp18 and Asp20 peptide fusion proteins. A375 cells were co-transfected with increasing amounts of the p300 gene (dark blue bars, 0 µg; brown bars, 1 µg; yellow bars, 2 µg and light blue bars, 5 µg), and fixed levels of p21-Luc (2 µg), β-Gal-reporter (1 µg) and the EGFP-peptide fusion vectors (100 ng), the cells were then processed for analysis of p53 activity as described in the legend for Figure 2C. (F) EGFP-Asp18 and Asp20 peptide fusion proteins inhibit p53-dependent transactivation in irradiated cells. EGFP-constructs (100 ng) were transiently transfected with 2 µg p21-Luc or 2 µg control-Luc and 1 µg control-β-Gal-reporter into A375 cells that were either untreated (dark blue bars), damaged with 20 J/m2 UV-C (brown bars), or with 5 Gy ionizing radiation (yellow bars). The cells were processed for analysis of p53 activity as described in the legend for Figure 2C.
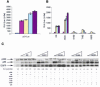
Fig. 3. In vivo inhibition of ectopically expressed p53 from the p21 promoter by phospho-peptide mimetic fusion proteins. (A) Stimulation of p53 activity by co-transfection with p300. Saos-2 cells were transiently co-transfected with 1 µg pCMV-p53, 2 µg p21-Luc, 1 µg pCMVβ-Gal and increasing amounts of pCMV-βp300: brown bars, 0 ng; yellow bars, 1 µg; light blue bars, 2 µg and purple bars, 5 µg. As a negative control, (dark blue bars) 5 µg pCMV-βp300, 2 µg p21-Luc and 1 µg pCMVβ-Gal were co-transfected. The cells were harvested 30 h post-transfection and the relative activity is expressed as a ratio of luciferase activity to β-Gal activity. (B) EGFP-S20D peptide inhibits p300 induction of p53-dependent gene expression. Saos-2 cells were transiently co-transfected with 1 µg pCMV-p53, 2 µg p21-Luc, 5 µg pCMVβp300, 1 µg pCMVβ-Gal and increasing amounts of EGFP-constructs as indicated (yellow bars, 1 µg; light blue bars, 2 µg and purple bars, 5 µg). To function as a critical control, 5 µg of each EGFP construct and 5 µg of pCMVβp300 were co-transfected with 2 µg p21-Luc and pCMVβGal (brown bars), and as a negative control (dark blue bars), 5 µg pCMV-βp300, 2 µg p21-Luc and 1 µg pCMVβ-Gal co-transfected. The cells were processed as described in the legend of Figure 3A. (C) Immunoblots of p53 and EGFP-fusion proteins in transfected Saos-2 cells. Lysates from transfected Saos-2 cells (as described in Figure 3B) were normalized for protein content by Bradford assay and loading for immunoblots was confirmed by Red Ponceau staining. The constructs transfected (in µg) are highlighted by the legend above the Figure (increasing amounts of EGFP fused to NS, BOX-I, S15D, T18D, and S20D BOX-I domain peptides) and are described below the Figure as ‘+’. The levels of p300, p53 and EGFP proteins are in the top, middle or bottom panels, respectively. p53-dependent activity (in RLUs) from Figure 3B is listed below the immunoblots for direct comparison of p53 activity to p53 protein and EGFP protein levels.
Similar articles
- DNA-dependent acetylation of p53 by the transcription coactivator p300.
Dornan D, Shimizu H, Perkins ND, Hupp TR. Dornan D, et al. J Biol Chem. 2003 Apr 11;278(15):13431-41. doi: 10.1074/jbc.M211460200. Epub 2002 Dec 23. J Biol Chem. 2003. PMID: 12499368 - Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers.
Craig AL, Burch L, Vojtesek B, Mikutowska J, Thompson A, Hupp TR. Craig AL, et al. Biochem J. 1999 Aug 15;342 ( Pt 1)(Pt 1):133-41. Biochem J. 1999. PMID: 10432310 Free PMC article. - Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2.
Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Unger T, et al. EMBO J. 1999 Apr 1;18(7):1805-14. doi: 10.1093/emboj/18.7.1805. EMBO J. 1999. PMID: 10202144 Free PMC article. - The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo.
Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, Hupp TR. Shimizu H, et al. J Biol Chem. 2002 Aug 9;277(32):28446-58. doi: 10.1074/jbc.M202296200. Epub 2002 Mar 29. J Biol Chem. 2002. PMID: 11925449 - Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface.
Chène P. Chène P. Mol Cancer Res. 2004 Jan;2(1):20-8. Mol Cancer Res. 2004. PMID: 14757842 Review.
Cited by
- Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2.
Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Ferreon JC, et al. Proc Natl Acad Sci U S A. 2009 Apr 21;106(16):6591-6. doi: 10.1073/pnas.0811023106. Epub 2009 Apr 8. Proc Natl Acad Sci U S A. 2009. PMID: 19357310 Free PMC article. - DeltaNp63 transcriptionally regulates ATM to control p53 Serine-15 phosphorylation.
Craig AL, Holcakova J, Finlan LE, Nekulova M, Hrstka R, Gueven N, DiRenzo J, Smith G, Hupp TR, Vojtesek B. Craig AL, et al. Mol Cancer. 2010 Jul 21;9:195. doi: 10.1186/1476-4598-9-195. Mol Cancer. 2010. PMID: 20663147 Free PMC article. - Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters.
Loughery J, Cox M, Smith LM, Meek DW. Loughery J, et al. Nucleic Acids Res. 2014 Jul;42(12):7666-80. doi: 10.1093/nar/gku501. Epub 2014 Jun 13. Nucleic Acids Res. 2014. PMID: 24928858 Free PMC article. - Tumour suppression by p53: a role for the DNA damage response?
Meek DW. Meek DW. Nat Rev Cancer. 2009 Oct;9(10):714-23. doi: 10.1038/nrc2716. Epub 2009 Sep 4. Nat Rev Cancer. 2009. PMID: 19730431 Review. - A function for the RING finger domain in the allosteric control of MDM2 conformation and activity.
Wawrzynow B, Pettersson S, Zylicz A, Bramham J, Worrall E, Hupp TR, Ball KL. Wawrzynow B, et al. J Biol Chem. 2009 Apr 24;284(17):11517-30. doi: 10.1074/jbc.M809294200. Epub 2009 Feb 2. J Biol Chem. 2009. PMID: 19188367 Free PMC article.
References
- Bottger A., Bottger, V., Sparks, A., Liu, W.L., Howard, S.F. and Lane, D.P. (1997) Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol., 7, 860–869. - PubMed
- Burch L.R., Midgley, C.A., Currie, R.A., Lane, D.P. and Hupp, T.R. (2000) Mdm2 binding to a conformationally sensitive domain on p53 can be modulated by RNA. FEBS Lett., 472, 93–98. - PubMed
- Craig A.L., Blaydes, J.P., Burch, L.R., Thompson, A.M. and Hupp, T.R. (1999a) Dephosphorylation of p53 at Ser20 after cellular exposure to low levels of non-ionizing radiation. Oncogene, 18, 6305–6312. - PubMed
- Craig A.L., Burch, L., Vojtesek, B., Mikutowska, J., Thompson, A. and Hupp, T.R. (1999b) Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem. J., 342, 133–141. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous