The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression - PubMed (original) (raw)
The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression
A Klochendler-Yeivin et al. EMBO Rep. 2000 Dec.
Abstract
The assembly of eukaryotic DNA into nucleosomes and derived higher order structures constitutes a barrier for transcription, replication and repair. A number of chromatin remodeling complexes, as well as histone acetylation, were shown to facilitate gene activation. To investigate the function of two closely related mammalian SWI/SNF complexes in vivo, we inactivated the murine SNF5/INI1 gene, a common subunit of these two complexes. Mice lacking SNF5 protein stop developing at the peri-implantation stage, showing that the SWI/SNF complex is essential for early development and viability of early embryonic cells. Furthermore, heterozygous mice develop nervous system and soft tissue sarcomas. In these tumors the wild-type allele was lost, providing further evidence that SNF5 functions as a tumor suppressor gene in certain cell types.
Figures
Fig. 1. Targeted disruption of the SNF5 gene. (A) Schematic representation of the gene targeting strategy. Exons are indicated by black boxes. The positions of 5′ and 3′ external probes are represented by gray boxes. Primers used for PCR analysis of embryos and mice are indicated by arrows. P, _Pst_I; X, _Xba_I; B, _Bam_HI; S, _Sac_I. (B) Southern blot analysis of genomic DNA from G418/gancyclovir-resistant ES clones. _Pst_I- and _Bam_HI-digested DNAs were hybridized with the 5′ and 3′ probes, respectively. One wild-type (+/+) and three heterozygous (+/–) clones are presented. The existence of a single integration site at the targeted locus was verified using _Bam_HI-digested DNA and a neo probe, giving a single 8.5 kb fragment (not shown).
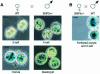
Fig. 2. Expression of the SNF5_–_lacZ allele in pre-implantation embryos. All cleavage stages were obtained from in vitro culture initiated at the one-cell stage. Fertilized oocytes were collected from mated superovulated females: (A) wild-type B6SJL F1 female × SNF5+/lacZ C57Bl6 × 129Sv male; (B) SNF5+/lacZ C57Bl6 × 129Sv female × wild-type C57Bl6 × 129Sv males. Recovery of oocytes and culture conditions were performed as described (Hogan, 1994). β-galactosidase activity was detected by whole mount staining with X-gal substrate. Scale bar, 20 µm.
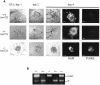
Fig. 3. Blastocyst outgrowth and apoptosis studies. (A) Day 3.5 p.c. embryos were isolated from SNF5+/lacZ intercrosses and cultured in 96-well plates for 4 days. The phase contrast view shows a growing inner cell mass (ICM) node and a single layer of trophoblastic giant cells (TGC) cells in wild-type and heterozygous blastocysts after 1, 2 and 4 days in culture, while homozygous mutants are impaired in both trophectoderm and ICM outgrowth. After 4 days of culture, TUNEL assay was performed. Fluorescein (TUNEL) and 4′-6-diamidine-2-phenylindole (DAPI) fluorescent images are shown. Scale bar, 60 µm. (B) Examples of PCR genotyping of cultured blastocysts.
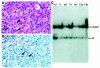
Fig. 4. SNF5 heterozygous mice are predisposed to cancer. (A) Microscopic features of tumors (5-month-old mouse; tumor from thoracic wall). Tumor cells appear as polygonal or elongated undifferentiated cells, with large clear pleomorphic nucleus, conspicuous nucleoli and eosinophilic cytoplasm. Some cells (arrows) display the classical appearance of rhabdoid cells: large excentric nucleus, with a prominent nucleolus, and hyalin-like inclusions in the cytoplasm. Hematoxylin–eosin staining, original magnification ×500. (B) Vimentin expression in tumor cells (5-month-old mouse; paravertebral tumor). Numerous cells are strongly positive and vimentin is often visible as perinuclear cytoplasmic spots (arrow). Anti-vimentin, hematoxylin counterstain, original magnification ×500. (C) Loss of heterozygosity (LOH) analysis. Southern blot analysis of four representative normal (N) and tumor (T) DNA samples prepared from SNF5+/– mice. Note the under-representation of the wild-type allele in the tumors; it remains detectable, probably due to contamination by surrounding normal cells. Tumor 1 (T1) was localized beneath the optic chiasma (7-month-old mouse); tumor 2 (T2) was a subcutaneous tumor of the cheek (7-month-old mouse); tumors 3a and 3b (T3a and T3b) developed in the thoracic and abdominal wall in a single 6-month-old animal. Control DNA samples were extracted from normal tissues close to the lesions.
Similar articles
- Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex.
Klochendler-Yeivin A, Picarsky E, Yaniv M. Klochendler-Yeivin A, et al. Mol Cell Biol. 2006 Apr;26(7):2661-74. doi: 10.1128/MCB.26.7.2661-2674.2006. Mol Cell Biol. 2006. PMID: 16537910 Free PMC article. - Integrase interactor 1 in health and disease.
Das S. Das S. Curr Protein Pept Sci. 2015;16(6):478-90. doi: 10.2174/1389203716666150316115156. Curr Protein Pept Sci. 2015. PMID: 25772157 Review. - Functional interaction of the retinoblastoma and Ini1/Snf5 tumor suppressors in cell growth and pituitary tumorigenesis.
Guidi CJ, Mudhasani R, Hoover K, Koff A, Leav I, Imbalzano AN, Jones SN. Guidi CJ, et al. Cancer Res. 2006 Aug 15;66(16):8076-82. doi: 10.1158/0008-5472.CAN-06-1451. Cancer Res. 2006. PMID: 16912184 - Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5.
Roberts CW, Leroux MM, Fleming MD, Orkin SH. Roberts CW, et al. Cancer Cell. 2002 Nov;2(5):415-25. doi: 10.1016/s1535-6108(02)00185-x. Cancer Cell. 2002. PMID: 12450796 - Chromatin remodeling: from transcription to cancer.
Yaniv M. Yaniv M. Cancer Genet. 2014 Sep;207(9):352-7. doi: 10.1016/j.cancergen.2014.03.006. Epub 2014 Mar 22. Cancer Genet. 2014. PMID: 24825771 Review.
Cited by
- Novel genomic prognostic biomarkers for dogs with cancer.
Chon E, Sakthikumar S, Tang M, Hamilton MJ, Vaughan A, Smith A, Sommer B, Robat C, Manley C, Mullin C, Ohashi E, Manor E, Custis J, Intile J, Shiu KB, Parshley L, Bergman N, Sheppard-Olivares S, Hafeman S, Wright Z, Haworth D, Hendricks W, Wang G. Chon E, et al. J Vet Intern Med. 2023 Nov-Dec;37(6):2410-2421. doi: 10.1111/jvim.16893. Epub 2023 Oct 6. J Vet Intern Med. 2023. PMID: 37801037 Free PMC article. - Atypical teratoid/rhabdoid tumour with CDK6 amplification in a child: a case report and literature review.
Li Z, Wang Y, Liu Y, Jiang Y, Han X, Zhao L, Li Y. Li Z, et al. Front Pediatr. 2023 Aug 22;11:1237572. doi: 10.3389/fped.2023.1237572. eCollection 2023. Front Pediatr. 2023. PMID: 37727617 Free PMC article. - SWI/SNF Complex Alterations in Tumors with Rhabdoid Features: Novel Therapeutic Approaches and Opportunities for Adoptive Cell Therapy.
Soto-Castillo JJ, Llavata-Marti L, Fort-Culillas R, Andreu-Cobo P, Moreno R, Codony C, García Del Muro X, Alemany R, Piulats JM, Martin-Liberal J. Soto-Castillo JJ, et al. Int J Mol Sci. 2023 Jul 6;24(13):11143. doi: 10.3390/ijms241311143. Int J Mol Sci. 2023. PMID: 37446319 Free PMC article. Review. - A chromatin remodelling SWI/SNF subunit, Snr1, regulates neural stem cell determination and differentiation.
Keegan SE, Haskins J, Simmonds AJ, Hughes SC. Keegan SE, et al. Development. 2023 Jul 1;150(13):dev201484. doi: 10.1242/dev.201484. Epub 2023 Jun 30. Development. 2023. PMID: 37294080 Free PMC article. - (mis)-Targeting of SWI/SNF complex(es) in cancer.
Reddy D, Bhattacharya S, Workman JL. Reddy D, et al. Cancer Metastasis Rev. 2023 Jun;42(2):455-470. doi: 10.1007/s10555-023-10102-5. Epub 2023 Apr 24. Cancer Metastasis Rev. 2023. PMID: 37093326 Free PMC article. Review.
References
- Bultman S., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Chambon, P., Crabtree, G. and Magnuson, T. (2000) A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell, in press. - PubMed
- Carlson M. and Laurent, B.C. (1994) The SWI/SNF family of global transcriptional activators. Curr. Opin. Cell Biol., 6, 396–402. - PubMed
- Dunaief J.L., Strober, B.E., Guha, S., Khavari, P.A., Alin, K., Luban, J., Begemann, M., Crabtree, G.R. and Goff, S.P. (1994) The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell, 79, 119–130. - PubMed
- Goodman R.H. and Smolik, S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. - PubMed
- Hogan B., Beddington, R., Costantini, F. and Lacy, E. (1994) Manipulating the Mouse Embryo. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases