Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli - PubMed (original) (raw)
Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli
M Sosio et al. Nucleic Acids Res. 2001.
Abstract
We developed a method for the reconstruction of a 100 kb DNA fragment into a bacterial artificial chromosome (BAC). The procedure makes use of iterative rounds of homologous recombination in Escherichia coli. Smaller, overlapping fragments of cloned DNA, such as cosmid clones, are required. They are transferred first into a temperature-sensitive replicon and then into the BAC of choice. We demonstrated the usefulness of this procedure by assembling a 90 kb genomic segment into an E.coli-STREPTOMYCES: artificial chromosome (ESAC). Using this procedure, ESACs are easy to handle and remarkably more stable than the starting cosmids.
Figures
Figure 1
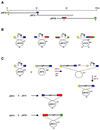
Starting genomic segment and constructs required for assembly. (A) The 90 kb P.rosea genomic fragment and the three cosmids pRP16, pRP31 and pRP58. Fragments A_–_D are color-coded. (B) The in vitro generated plasmids pMAB1, pMBC1, pMCD1 and pPAD1. (C) Scheme of cointegrate formation and resolution between each cosmid and the cognate ts construct. The interrupted empty bars designate the genomic segments comprised between fragments A and B, B and C, and C and D. Chloramphenicol and kanamycin are abbreviated to Cm and Km, respectively. cat, aph and tet represent Cm, Km and tetracycline resistance genes, respectively; ts, the temperature-sensitive replication origin. _Dra_I sites in pPAD1 are indicated as D.
Figure 2
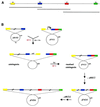
Strategy for assembly into an ESAC. (A) A genomic segment, covered by three overlapping cosmids, with the positions of fragments A_–_D. (B) Schematic of the recombination steps. The first crossover event between pMAB2 and pPAD1 leads to formation of the cointegrate (after recombination via fragment A), while the second crossover event (via fragment B) leads to the resolved cointegrate. The starting pPAC-S1 derivative is then enlarged in a step-wise fashion by cointegrate formation and resolution with the appropriate ts plasmids. Fragments A_–_D are color-coded as in Figure 1. Abbreviations are as in Figure 1.
Figure 3
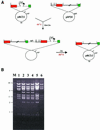
Isolation and resolution of a cointegrate. (A) Selection for the pMCD1::pRP58 cointegrate at 44°C (non-permissive temperature) and resolution at 30°C to yield pMCD2. The antibiotics used for selection are indicated. _Bam_HI sites are indicated by bars. Other symbols and abbreviations are as in Figure 1. (B) Analysis of cointegrates and resolved cointegrates. The _Bam_HI profiles of pMCD2 (lane 1), of three independent pMCD1::pRP58 cointegrates (lanes 2–4) and of pRP58 (lanes 5–6) are shown. M, molecular weight marker, with relevant sizes (in kb) on the left.
Figure 4
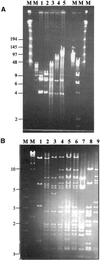
Analysis of the growing ESACs. (A) Pulse field gel electrophoresis analysis of ESACs after _Dra_I digestion: pPAC-S1 (lane 1), pPAD1 (lane 2), PAD2 (lane 3), PAD4 (lane 4) and PAD6 (lane 5). M, molecular weight markers, with relevant sizes (in kb) on the left. Running conditions: 1% agarose gel in 0.5× TBE, 6–9 V/cm for 20 h at 14°C. (B) Comparisons of the _Bam_HI profiles. Lanes 1–6 contain pPAD1, PAD2, PAD3, PAD4, PAD5 and PAD6, respectively; pRP16, pRP31 and pRP58 are in lanes 7–9, respectively. M, molecular weight marker, with relevant sizes (in kb) on the left.
Similar articles
- Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors.
Tao Q, Zhang HB. Tao Q, et al. Nucleic Acids Res. 1998 Nov 1;26(21):4901-9. doi: 10.1093/nar/26.21.4901. Nucleic Acids Res. 1998. PMID: 9776751 Free PMC article. - Transposon and transposome mutagenesis of plasmids, cosmids, and BACs.
McGregor A. McGregor A. Methods Mol Biol. 2003;235:233-45. doi: 10.1385/1-59259-409-3:233. Methods Mol Biol. 2003. PMID: 12904666 No abstract available. - Enhanced efficiency of pSV1-RecA-based BAC recombineering.
Cox DH, Carvajal JJ, Rigby PW. Cox DH, et al. Biotechniques. 2002 Dec;33(6):1206-8. doi: 10.2144/02336bm04. Biotechniques. 2002. PMID: 12503299 No abstract available. - ET-cloning: think recombination first.
Muyrers JP, Zhang Y, Stewart AF. Muyrers JP, et al. Genet Eng (N Y). 2000;22:77-98. doi: 10.1007/978-1-4615-4199-8_6. Genet Eng (N Y). 2000. PMID: 11501382 Review. No abstract available. - Recombineering: a powerful new tool for mouse functional genomics.
Copeland NG, Jenkins NA, Court DL. Copeland NG, et al. Nat Rev Genet. 2001 Oct;2(10):769-79. doi: 10.1038/35093556. Nat Rev Genet. 2001. PMID: 11584293 Review.
Cited by
- Biosynthesis of Aurodox, a Type III Secretion System Inhibitor from Streptomyces goldiniensis.
McHugh RE, Munnoch JT, Braes RE, McKean IJW, Giard J, Taladriz-Sender A, Peschke F, Burley GA, Roe AJ, Hoskisson PA. McHugh RE, et al. Appl Environ Microbiol. 2022 Aug 9;88(15):e0069222. doi: 10.1128/aem.00692-22. Epub 2022 Jul 18. Appl Environ Microbiol. 2022. PMID: 35867559 Free PMC article. - Artificial chromosomes to explore and to exploit biosynthetic capabilities of actinomycetes.
Alduina R, Gallo G. Alduina R, et al. J Biomed Biotechnol. 2012;2012:462049. doi: 10.1155/2012/462049. Epub 2012 Aug 7. J Biomed Biotechnol. 2012. PMID: 22919271 Free PMC article. Review. - The Saccharomyces cerevisiae SCRaMbLE system and genome minimization.
Dymond J, Boeke J. Dymond J, et al. Bioeng Bugs. 2012 May-Jun;3(3):168-71. doi: 10.4161/bbug.19543. Epub 2012 May 1. Bioeng Bugs. 2012. PMID: 22572789 Free PMC article. - The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences.
Gao H, Bhattacharyya MK. Gao H, et al. BMC Plant Biol. 2008 Mar 19;8:29. doi: 10.1186/1471-2229-8-29. BMC Plant Biol. 2008. PMID: 18366691 Free PMC article. - Novel bacterial artificial chromosome vector pUvBBAC for use in studies of the functional genomics of Listeria spp.
Hain T, Otten S, von Both U, Chatterjee SS, Technow U, Billion A, Ghai R, Mohamed W, Domann E, Chakraborty T. Hain T, et al. Appl Environ Microbiol. 2008 Mar;74(6):1892-901. doi: 10.1128/AEM.00415-07. Epub 2008 Jan 25. Appl Environ Microbiol. 2008. PMID: 18223114 Free PMC article.
References
- Ioannou P.A., Amemiya,C.T., Garnes,J., Kroisel,P.M., Shizuya,H., Chen,C., Batzer,M.A. and de Jong,P.J. (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat. Genet., 6, 84–89. - PubMed
- Groisman E.A. and Ochman,H. (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell, 87, 791–794. - PubMed
- Martìn J.F. and Liras,P. (1989) Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol., 43, 173–206. - PubMed
- Yang X.W., Model,P. and Heintz,N. (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol., 15, 859–865. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources