Naked cuticle targets dishevelled to antagonize Wnt signal transduction - PubMed (original) (raw)
Naked cuticle targets dishevelled to antagonize Wnt signal transduction
R Rousset et al. Genes Dev. 2001.
Abstract
In Drosophila embryos the protein Naked cuticle (Nkd) limits the effects of the Wnt signal Wingless (Wg) during early segmentation. nkd loss of function results in segment polarity defects and embryonic death, but how nkd affects Wnt signaling is unknown. Using ectopic expression, we find that Nkd affects, in a cell-autonomous manner, a transduction step between the Wnt signaling components Dishevelled (Dsh) and Zeste-white 3 kinase (Zw3). Zw3 is essential for repressing Wg target-gene transcription in the absence of a Wg signal, and the role of Wg is to relieve this inhibition. Our double-mutant analysis shows that, in contrast to Zw3, Nkd acts when the Wg pathway is active to restrain signal transduction. Yeast two hybrid and in vitro experiments indicate that Nkd directly binds to the basic-PDZ region of Dsh. Specially timed Nkd overexpression is capable of abolishing Dsh function in a distinct signaling pathway that controls planar-cell polarity. Our results suggest that Nkd acts directly through Dsh to limit Wg activity and thus determines how efficiently Wnt signals stabilize Armadillo (Arm)/beta-catenin and activate downstream genes.
Figures
Figure 1
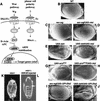
Epistasis study in the eye and embryo. (A) Schematic diagram of the Wg pathway and the planar cell polarity pathway. Arrows and bars show positive and negative actions, respectively. P represents the phosphorylated state of Dsh. See text for details. (B–J) Nkd can suppress Wg and Dsh misexpression eye phenotypes, but not the _ArmS10_misexpression eye phenotype. Ventral is to the left and anterior up. (B) The wild-type adult eye consists of an array of ommatidia and interommatidial bristles. (C,E) The sev-wg or UAS-dsh eyes lack bristles and/or ommatidia. (G) The UAS-armS10 eye has disrupted ommatidia and loss of bristles (average number of bristles/eye = 11.6; n = 8). (D,F) Co-misexpression of UAS-nkd dramatically suppressed the sev-wg (n = 8) and the UAS-dsh (n = 8) eye phenotypes. (H) Co-misexpression of UAS-nkd did not suppress the UAS-armS10 phenotype (average number of bristles/eye = 5.4; n = 8). UAS-GPI-Dfz2 suppresses the sev-wg loss of bristle phenotype (I) but does not suppress the UAS-dsh phenotype (J). (K,L) Injection of _Xenopus GSK3_β mRNA into nkd mutant embryos can restore denticles to the nkd embryos. Anterior is up. (K) nkd mutant embryos lack ventral denticle belts. (L) Injection of _GSK3_β into nkd7H16 embryos restored ventral denticles (brackets). nkd7H16 mutant embryos were identified by a Ubx mutation resulting in the transformation of the first abdominal segment A1 to the third thoracic segment T3 (arrow).
Figure 2
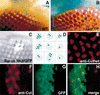
nkd autonomously regulates interommatidial bristle formation. (A) Wild-type eye margin at dorsum of head shows bristle suppression 0–1 ommatidial rows (blue brackets) from eye margin. (B) nkd7E89 mutant eye shows suppression of bristles 3–5 ommatidial rows from eye margin. Occasional bristles are present closer to eye margin (yellow arrow). No bristle phenotype was seen with the weaker nkd9G33 allele, whereas h1 nkd7H16 eyes are small and rough (not shown), possibly due to h/nkd interactions (Zeng et al. 2000) and hence could not be scored for this phenotype. (C,D) Nkd/GFP misexpression clones in sev-wg adult eyes. (C) Bristles are restored only in the region of the clone marked by GFP. (D) Cartoon depiction of nine adult eye clones (green color); vertical black lines represent bristles. (E) Cut nuclear localization in bristle cells of a wild-type pupal eye disc. (F,G) Restoration of bristle cells in a sev-wg pupal eye, revealed by Cut localization (red color), is confined to the region of the Nkd/GFP misexpression clone (green color); the clone begins at the edge of the disc (left) and continues inward (right). (H) Merged image of F and G shows that GFP and Cut colocalize in bristle cell precursor nuclei present in the clone.
Figure 3
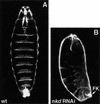
Cuticle phenotypes of embryos injected with nkd dsRNA. (A) Wild-type (wt) embryo shortly before hatching. The ventral side is characterized by denticle belts separated with naked cuticle. (B) nkd RNAi mimics the nkd phenotype. Embryos are shorter and lack denticle belts, resembling _nkd_−/− embryos (see Fig. 1K). nkd RNAi embryos also show the characteristic defects in the Filzkörper (FK).
Figure 4
Nkd and Dsh directly interact in the yeast two-hybrid system, coimmunoprecipitation and GST pull-down assays. (A) Interaction between Nkd and Dsh in the yeast two-hybrid system using a liquid culture β-galactosidase assay. Yeast cells were cotransformed with plasmids expressing the GAL4 DNA-binding domain (GB, amino acids [aa] 1–147) either alone (−) or in fusion with Nkd, along with plasmids expressing the GAL4 activation domain (GAD, aa 768–881) alone (−) or in fusion with Dsh. The interaction between GB-Nkd and GAD-Zw3, GAD-Arm, or GAD-Dfz2CT (intracellular portion of Dfz2) was also tested. Murine p53 (aa 72–390) fused to GB and SV40 large T-antigen (T-Ag, aa 87–708) fused to GAD were used as positive controls. For each transformation, results corresponding to the mean of 2-independent yeast colonies are shown. The values are expressed in Miller units. The activation was confirmed using a second reporter gene (ADE2) present in the yeast strain and protein blots of yeast extracts showed that the different fusion proteins accumulated to similar levels (data not shown). (B) Coimmunoprecipitation of Nkd and Dsh in COS-7 cells. Extracts from COS-7 cells expressing Nkd-myc and Dsh-myc were subjected to coimmunoprecipitation with anti-Dsh antibody (α Dsh Ab) (+). As a control, the anti-Dsh antibody was omitted (−). The eluted protein from the beads (P) and one-tenth of the supernatant (S), as well as proteins corresponding to one-tenth of the input (I) were analyzed by Western blot using anti-c-Myc antibody. (C,D) Interaction between Nkd and Dsh in the GST pull-down assay. Bacterially expressed GST-Nkd protein was incubated with [35S]methionine-Dsh, produced and labeled by in vitro translation (C), whereas GST-Dsh was incubated with [35S]methionine-Nkd (D). As controls, GST-Nkd or GST-Dsh proteins were also tested for their interaction with [35S]methionine-Luciferase (Luc). The eluted proteins from the beads (P) and one-tenth of the supernatant (S) were separated by SDS-PAGE and viewed using a phosphorimager (Molecular Dynamics).
Figure 4
Nkd and Dsh directly interact in the yeast two-hybrid system, coimmunoprecipitation and GST pull-down assays. (A) Interaction between Nkd and Dsh in the yeast two-hybrid system using a liquid culture β-galactosidase assay. Yeast cells were cotransformed with plasmids expressing the GAL4 DNA-binding domain (GB, amino acids [aa] 1–147) either alone (−) or in fusion with Nkd, along with plasmids expressing the GAL4 activation domain (GAD, aa 768–881) alone (−) or in fusion with Dsh. The interaction between GB-Nkd and GAD-Zw3, GAD-Arm, or GAD-Dfz2CT (intracellular portion of Dfz2) was also tested. Murine p53 (aa 72–390) fused to GB and SV40 large T-antigen (T-Ag, aa 87–708) fused to GAD were used as positive controls. For each transformation, results corresponding to the mean of 2-independent yeast colonies are shown. The values are expressed in Miller units. The activation was confirmed using a second reporter gene (ADE2) present in the yeast strain and protein blots of yeast extracts showed that the different fusion proteins accumulated to similar levels (data not shown). (B) Coimmunoprecipitation of Nkd and Dsh in COS-7 cells. Extracts from COS-7 cells expressing Nkd-myc and Dsh-myc were subjected to coimmunoprecipitation with anti-Dsh antibody (α Dsh Ab) (+). As a control, the anti-Dsh antibody was omitted (−). The eluted protein from the beads (P) and one-tenth of the supernatant (S), as well as proteins corresponding to one-tenth of the input (I) were analyzed by Western blot using anti-c-Myc antibody. (C,D) Interaction between Nkd and Dsh in the GST pull-down assay. Bacterially expressed GST-Nkd protein was incubated with [35S]methionine-Dsh, produced and labeled by in vitro translation (C), whereas GST-Dsh was incubated with [35S]methionine-Nkd (D). As controls, GST-Nkd or GST-Dsh proteins were also tested for their interaction with [35S]methionine-Luciferase (Luc). The eluted proteins from the beads (P) and one-tenth of the supernatant (S) were separated by SDS-PAGE and viewed using a phosphorimager (Molecular Dynamics).
Figure 4
Nkd and Dsh directly interact in the yeast two-hybrid system, coimmunoprecipitation and GST pull-down assays. (A) Interaction between Nkd and Dsh in the yeast two-hybrid system using a liquid culture β-galactosidase assay. Yeast cells were cotransformed with plasmids expressing the GAL4 DNA-binding domain (GB, amino acids [aa] 1–147) either alone (−) or in fusion with Nkd, along with plasmids expressing the GAL4 activation domain (GAD, aa 768–881) alone (−) or in fusion with Dsh. The interaction between GB-Nkd and GAD-Zw3, GAD-Arm, or GAD-Dfz2CT (intracellular portion of Dfz2) was also tested. Murine p53 (aa 72–390) fused to GB and SV40 large T-antigen (T-Ag, aa 87–708) fused to GAD were used as positive controls. For each transformation, results corresponding to the mean of 2-independent yeast colonies are shown. The values are expressed in Miller units. The activation was confirmed using a second reporter gene (ADE2) present in the yeast strain and protein blots of yeast extracts showed that the different fusion proteins accumulated to similar levels (data not shown). (B) Coimmunoprecipitation of Nkd and Dsh in COS-7 cells. Extracts from COS-7 cells expressing Nkd-myc and Dsh-myc were subjected to coimmunoprecipitation with anti-Dsh antibody (α Dsh Ab) (+). As a control, the anti-Dsh antibody was omitted (−). The eluted protein from the beads (P) and one-tenth of the supernatant (S), as well as proteins corresponding to one-tenth of the input (I) were analyzed by Western blot using anti-c-Myc antibody. (C,D) Interaction between Nkd and Dsh in the GST pull-down assay. Bacterially expressed GST-Nkd protein was incubated with [35S]methionine-Dsh, produced and labeled by in vitro translation (C), whereas GST-Dsh was incubated with [35S]methionine-Nkd (D). As controls, GST-Nkd or GST-Dsh proteins were also tested for their interaction with [35S]methionine-Luciferase (Luc). The eluted proteins from the beads (P) and one-tenth of the supernatant (S) were separated by SDS-PAGE and viewed using a phosphorimager (Molecular Dynamics).
Figure 4
Nkd and Dsh directly interact in the yeast two-hybrid system, coimmunoprecipitation and GST pull-down assays. (A) Interaction between Nkd and Dsh in the yeast two-hybrid system using a liquid culture β-galactosidase assay. Yeast cells were cotransformed with plasmids expressing the GAL4 DNA-binding domain (GB, amino acids [aa] 1–147) either alone (−) or in fusion with Nkd, along with plasmids expressing the GAL4 activation domain (GAD, aa 768–881) alone (−) or in fusion with Dsh. The interaction between GB-Nkd and GAD-Zw3, GAD-Arm, or GAD-Dfz2CT (intracellular portion of Dfz2) was also tested. Murine p53 (aa 72–390) fused to GB and SV40 large T-antigen (T-Ag, aa 87–708) fused to GAD were used as positive controls. For each transformation, results corresponding to the mean of 2-independent yeast colonies are shown. The values are expressed in Miller units. The activation was confirmed using a second reporter gene (ADE2) present in the yeast strain and protein blots of yeast extracts showed that the different fusion proteins accumulated to similar levels (data not shown). (B) Coimmunoprecipitation of Nkd and Dsh in COS-7 cells. Extracts from COS-7 cells expressing Nkd-myc and Dsh-myc were subjected to coimmunoprecipitation with anti-Dsh antibody (α Dsh Ab) (+). As a control, the anti-Dsh antibody was omitted (−). The eluted protein from the beads (P) and one-tenth of the supernatant (S), as well as proteins corresponding to one-tenth of the input (I) were analyzed by Western blot using anti-c-Myc antibody. (C,D) Interaction between Nkd and Dsh in the GST pull-down assay. Bacterially expressed GST-Nkd protein was incubated with [35S]methionine-Dsh, produced and labeled by in vitro translation (C), whereas GST-Dsh was incubated with [35S]methionine-Nkd (D). As controls, GST-Nkd or GST-Dsh proteins were also tested for their interaction with [35S]methionine-Luciferase (Luc). The eluted proteins from the beads (P) and one-tenth of the supernatant (S) were separated by SDS-PAGE and viewed using a phosphorimager (Molecular Dynamics).
Figure 5
Nkd interacts with the basic/PDZ region of Dsh. (A) Interaction of different Dsh deletion mutants, fused to GAD, with GB or GB-Nkd in the yeast two-hybrid system. Yeast growth was evaluated using the ADE reporter gene present in the strain: Yeast colonies expressing the different combinations of fusion proteins, as indicated, were grown on medium containing (+) or lacking (−) adenine. (B,C) Using the GST pull-down assay, [35S]methionine-Dsh D1 to D6 were tested for their capacity to bind GST-Nkd (B), whereas in (C), GST-DshD6PDZ and GST-DshPDZ proteins were incubated with [35S]methionine-Nkd. The curved bands observed with D1 and D6 are due to comigration with a byproduct of GST-Nkd protein (data not shown). (D) Summary of domain mapping results from the yeast two-hybrid system (Y2H) and the GST pull-down assay (GST). (+) Positive interaction, (−) no interaction, (+/−) weak interaction, (n.d.) not determined. The different domains of Dsh, DIX, basic region (b), PDZ, and DEP, are indicated.
Figure 5
Nkd interacts with the basic/PDZ region of Dsh. (A) Interaction of different Dsh deletion mutants, fused to GAD, with GB or GB-Nkd in the yeast two-hybrid system. Yeast growth was evaluated using the ADE reporter gene present in the strain: Yeast colonies expressing the different combinations of fusion proteins, as indicated, were grown on medium containing (+) or lacking (−) adenine. (B,C) Using the GST pull-down assay, [35S]methionine-Dsh D1 to D6 were tested for their capacity to bind GST-Nkd (B), whereas in (C), GST-DshD6PDZ and GST-DshPDZ proteins were incubated with [35S]methionine-Nkd. The curved bands observed with D1 and D6 are due to comigration with a byproduct of GST-Nkd protein (data not shown). (D) Summary of domain mapping results from the yeast two-hybrid system (Y2H) and the GST pull-down assay (GST). (+) Positive interaction, (−) no interaction, (+/−) weak interaction, (n.d.) not determined. The different domains of Dsh, DIX, basic region (b), PDZ, and DEP, are indicated.
Figure 5
Nkd interacts with the basic/PDZ region of Dsh. (A) Interaction of different Dsh deletion mutants, fused to GAD, with GB or GB-Nkd in the yeast two-hybrid system. Yeast growth was evaluated using the ADE reporter gene present in the strain: Yeast colonies expressing the different combinations of fusion proteins, as indicated, were grown on medium containing (+) or lacking (−) adenine. (B,C) Using the GST pull-down assay, [35S]methionine-Dsh D1 to D6 were tested for their capacity to bind GST-Nkd (B), whereas in (C), GST-DshD6PDZ and GST-DshPDZ proteins were incubated with [35S]methionine-Nkd. The curved bands observed with D1 and D6 are due to comigration with a byproduct of GST-Nkd protein (data not shown). (D) Summary of domain mapping results from the yeast two-hybrid system (Y2H) and the GST pull-down assay (GST). (+) Positive interaction, (−) no interaction, (+/−) weak interaction, (n.d.) not determined. The different domains of Dsh, DIX, basic region (b), PDZ, and DEP, are indicated.
Figure 5
Nkd interacts with the basic/PDZ region of Dsh. (A) Interaction of different Dsh deletion mutants, fused to GAD, with GB or GB-Nkd in the yeast two-hybrid system. Yeast growth was evaluated using the ADE reporter gene present in the strain: Yeast colonies expressing the different combinations of fusion proteins, as indicated, were grown on medium containing (+) or lacking (−) adenine. (B,C) Using the GST pull-down assay, [35S]methionine-Dsh D1 to D6 were tested for their capacity to bind GST-Nkd (B), whereas in (C), GST-DshD6PDZ and GST-DshPDZ proteins were incubated with [35S]methionine-Nkd. The curved bands observed with D1 and D6 are due to comigration with a byproduct of GST-Nkd protein (data not shown). (D) Summary of domain mapping results from the yeast two-hybrid system (Y2H) and the GST pull-down assay (GST). (+) Positive interaction, (−) no interaction, (+/−) weak interaction, (n.d.) not determined. The different domains of Dsh, DIX, basic region (b), PDZ, and DEP, are indicated.
Figure 6
Effects of overproduced Nkd on planar cell polarity. (A) Wild-type (WT) wing pattern in region distal to posterior cross vein; same area shown in B–E. (B) Effect of overproduction of Nkd, which is similar to the phenotype shown in C, but different to the phenotypes shown in D and E. (C) Phenotype of loss of dsh function (dsh1 allele). (D) Phenotype of loss of frizzled function (fzJ22 allele). (E) Phenotype of loss of prickle function (pk30 allele). Arrows indicate hair orientation. (F) Wild-type wing pattern in the area shown in G and H: region posterior to vein 5. (G) Phenotype of heat shock promoter-driven frizzled expression. (H) Phenotype of heat shock promoter–driven fz expression in the presence of UAS-nkd. Almost complete suppression of polarity defects by Nkd overexpression is observed. High magnification shows that fz overexpression also induces double hair cells (G inset) and that their number decreases in the presence of Nkd (H inset).
Similar articles
- Vertebrate proteins related to Drosophila Naked Cuticle bind Dishevelled and antagonize Wnt signaling.
Wharton KA Jr, Zimmermann G, Rousset R, Scott MP. Wharton KA Jr, et al. Dev Biol. 2001 Jun 1;234(1):93-106. doi: 10.1006/dbio.2001.0238. Dev Biol. 2001. PMID: 11356022 - An unconventional nuclear localization motif is crucial for function of the Drosophila Wnt/wingless antagonist Naked cuticle.
Waldrop S, Chan CC, Cagatay T, Zhang S, Rousset R, Mack J, Zeng W, Fish M, Zhang M, Amanai M, Wharton KA Jr. Waldrop S, et al. Genetics. 2006 Sep;174(1):331-48. doi: 10.1534/genetics.106.061853. Epub 2006 Jul 18. Genetics. 2006. PMID: 16849595 Free PMC article. - Zinc-dependent interaction between dishevelled and the Drosophila Wnt antagonist naked cuticle.
Rousset R, Wharton KA Jr, Zimmermann G, Scott MP. Rousset R, et al. J Biol Chem. 2002 Dec 13;277(50):49019-26. doi: 10.1074/jbc.M203246200. Epub 2002 Sep 26. J Biol Chem. 2002. PMID: 12354775 - New steps in the Wnt/beta-catenin signal transduction pathway.
Sakanaka C, Sun TQ, Williams LT. Sakanaka C, et al. Recent Prog Horm Res. 2000;55:225-36. Recent Prog Horm Res. 2000. PMID: 11036939 Review. - Signaling through beta-catenin and Lef/Tcf.
Novak A, Dedhar S. Novak A, et al. Cell Mol Life Sci. 1999 Oct 30;56(5-6):523-37. doi: 10.1007/s000180050449. Cell Mol Life Sci. 1999. PMID: 11212302 Free PMC article. Review.
Cited by
- Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells.
Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Bao R, et al. PLoS One. 2012;7(11):e48670. doi: 10.1371/journal.pone.0048670. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144924 Free PMC article. - Nkd2 promotes the differentiation of dental follicle stem/progenitor cells into osteoblasts.
Chen C, Zhang J, Ling J, Du Y, Hou Y. Chen C, et al. Int J Mol Med. 2018 Nov;42(5):2403-2414. doi: 10.3892/ijmm.2018.3822. Epub 2018 Aug 14. Int J Mol Med. 2018. PMID: 30106129 Free PMC article. - Differentially expressed genes in embryonic cardiac tissues of mice lacking Folr1 gene activity.
Zhu H, Cabrera RM, Wlodarczyk BJ, Bozinov D, Wang D, Schwartz RJ, Finnell RH. Zhu H, et al. BMC Dev Biol. 2007 Nov 20;7:128. doi: 10.1186/1471-213X-7-128. BMC Dev Biol. 2007. PMID: 18028541 Free PMC article. - The Sno oncogene antagonizes Wingless signaling during wing development in Drosophila.
Quijano JC, Stinchfield MJ, Zerlanko B, Gibbens YY, Takaesu NT, Hyman-Walsh C, Wotton D, Newfeld SJ. Quijano JC, et al. PLoS One. 2010 Jul 16;5(7):e11619. doi: 10.1371/journal.pone.0011619. PLoS One. 2010. PMID: 20661280 Free PMC article. - Casein kinase Iepsilon modulates the signaling specificities of dishevelled.
Cong F, Schweizer L, Varmus H. Cong F, et al. Mol Cell Biol. 2004 Mar;24(5):2000-11. doi: 10.1128/MCB.24.5.2000-2011.2004. Mol Cell Biol. 2004. PMID: 14966280 Free PMC article.
References
- Ahmed Y, Hayashi S, Levine A, Wieschaus E. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell. 1998;93:1171–1182. - PubMed
- Bejsovec A, Wieschaus E. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development. 1993;119:501–517. - PubMed
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases