Fibroblast growth factors - PubMed (original) (raw)
Review
Fibroblast growth factors
D M Ornitz et al. Genome Biol. 2001.
Abstract
Fibroblast growth factors (FGFs) make up a large family of polypeptide growth factors that are found in organisms ranging from nematodes to humans. In vertebrates, the 22 members of the FGF family range in molecular mass from 17 to 34 kDa and share 13-71% amino acid identity. Between vertebrate species, FGFs are highly conserved in both gene structure and amino-acid sequence. FGFs have a high affinity for heparan sulfate proteoglycans and require heparan sulfate to activate one of four cell-surface FGF receptors. During embryonic development, FGFs have diverse roles in regulating cell proliferation, migration and differentiation. In the adult organism, FGFs are homeostatic factors and function in tissue repair and response to injury. When inappropriately expressed, some FGFs can contribute to the pathogenesis of cancer. A subset of the FGF family, expressed in adult tissue, is important for neuronal signal transduction in the central and peripheral nervous systems.
Figures
Figure 1
Gene structure of selected members of the Fgf family. Only the portion of each gene containing coding exons is shown. Constitutively expressed exons are in black; alternatively spliced exons are in gray. Fgfs1, 2, 4 and 9 contain the prototypic three-exon organization. For Fgf1, 5' untranslated exons are not shown; inclusion of these exons extends the gene by approximately 69 kb [78]. Fgf8 is an example of a gene with 5' alternative splicing, and Fgf13 demonstrates alternatively used 5' exons separated by over 30 kb. References: Fgf1 [78]; Fgf2 [79]; Fgf4 [80]; Fgf8 [52]; Fgf9 [81]; Fgf13 [76].
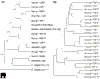
Figure 2
Evolutionary relationships within the FGF family. (a) Apparent evolutionary relationships between FGFs from vertebrates, invertebrates and a virus. Amino-acid sequences of nine representative FGFs were chosen from human and compared with FGFs from Drosophila, C. elegans, zebrafish and Autographa californica nuclear polyhedrosis virus. (b) Apparent evolutionary relationships of the 22 known human and murine FGFs. Sequences were aligned using Genetyxsequence analysis software and trees were constructed from the alignments using the neighbor-joining method.
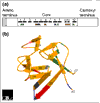
Figure 3
(a) Structural features of the FGF polypeptide. The amino terminus of some FGFs contains a signal sequence (shaded). All FGFs contain a core region that contains conserved amino-acid residues and conserved structural motifs. The locations of β strands within the core region are numbered and shown as black boxes. The heparin-binding region (pink) includes residues in the loop between β strands 1 and 2 and in β strands 10 and 11. Residues that contact the FGFR are shown in green (the region contacting Ig-domain 2 of the receptor), blue (contacting Ig-domain 3) and red (contacting the alternatively spliced region of Ig-domain 3). Amino-acid residues that contact the linker region are shown in gray [20]. (b) Three-dimensional structure of FGF2, a prototypical member of the FGF family. A ribbon diagram of FGF2 is shown; β strands are labeled 1-12 and regions of contact with the FGFR and heparin are color-coded as in (a) [22,24]. Image provided by M. Mohammadi.
Similar articles
- Signaling, internalization, and intracellular activity of fibroblast growth factor.
Wiedłocha A, Sørensen V. Wiedłocha A, et al. Curr Top Microbiol Immunol. 2004;286:45-79. doi: 10.1007/978-3-540-69494-6_3. Curr Top Microbiol Immunol. 2004. PMID: 15645710 Review. - Common and specific determinants for fibroblast growth factors in the ectodomain of the receptor kinase complex.
Wang F, Lu W, McKeehan K, Mohamedali K, Gabriel JL, Kan M, McKeehan WL. Wang F, et al. Biochemistry. 1999 Jan 5;38(1):160-71. doi: 10.1021/bi981758m. Biochemistry. 1999. PMID: 9890894 - Fibroblast (heparin-binding) growing factors in neuronal development and repair.
Haynes LW. Haynes LW. Mol Neurobiol. 1988 Winter;2(4):263-89. doi: 10.1007/BF02935635. Mol Neurobiol. 1988. PMID: 2855976 Review. - FGFs, heparan sulfate and FGFRs: complex interactions essential for development.
Ornitz DM. Ornitz DM. Bioessays. 2000 Feb;22(2):108-12. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. Bioessays. 2000. PMID: 10655030 Review.
Cited by
- Treatment of Oroantral Communication and Fistulas with the Use of Blood-Derived Platelet-Rich Preparations Rich in Growth Factors: A Systematic Review.
Adamska P, Kaczoruk-Wieremczuk M, Pylińska-Dąbrowska D, Stasiak M, Bartmański M, Zedler A, Studniarek M. Adamska P, et al. Int J Mol Sci. 2024 Oct 26;25(21):11507. doi: 10.3390/ijms252111507. Int J Mol Sci. 2024. PMID: 39519060 Free PMC article. Review. - The Immobilization of an FGF2-Derived Peptide on Culture Plates Improves the Production and Therapeutic Potential of Extracellular Vesicles from Wharton's Jelly Mesenchymal Stem Cells.
Lee Y, Lim KM, Bong H, Lee SB, Jeon TI, Lee SY, Park HS, Kim JY, Song K, Kang GH, Kim SJ, Song M, Cho SG. Lee Y, et al. Int J Mol Sci. 2024 Oct 4;25(19):10709. doi: 10.3390/ijms251910709. Int J Mol Sci. 2024. PMID: 39409038 Free PMC article. - FGF2 promotes the expansion of parietal mesothelial progenitor pools and inhibits BMP4-mediated smooth muscle cell differentiation.
Hwang Y, Shimamura Y, Tanaka J, Miura A, Sawada A, Sarmah H, Shimizu D, Kondo Y, Lee H, Martini F, Ninish Z, Yan KS, Yamada K, Mori M. Hwang Y, et al. Front Cell Dev Biol. 2024 Sep 23;12:1387237. doi: 10.3389/fcell.2024.1387237. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39376629 Free PMC article. - Exploring endocrine FGFs - structures, functions and biomedical applications.
Phan P, Ternier G, Edirisinghe O, Kumar TKS. Phan P, et al. Int J Biochem Mol Biol. 2024 Aug 25;15(4):68-99. doi: 10.62347/PALK2137. eCollection 2024. Int J Biochem Mol Biol. 2024. PMID: 39309613 Free PMC article. Review. - Recent developments in receptor tyrosine kinase inhibitors: A promising mainstay in targeted cancer therapy.
Kumar R, Goel H, Solanki R, Rawat L, Tabasum S, Tanwar P, Pal S, Sabarwal A. Kumar R, et al. Med Drug Discov. 2024 Sep;23:100195. doi: 10.1016/j.medidd.2024.100195. Epub 2024 Jul 1. Med Drug Discov. 2024. PMID: 39281823 Free PMC article.
References
- Kiefer P, Acland P, Pappin D, Peters G, Dickson C. Competition between nuclear localization and secretory signals determines the subcellular fate of a single CUG-initiated form of FGF3. EMBO J. 1994;13:4126–4136. This is one of first papers to show that an FGF can initiate from an upstream CUG codon and can be localized in the nucleus. - PMC - PubMed
- Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol. 1999;19:505–514. Identification of a high-molecular-weight form of FGF2 that initiates from an upsteam CUG codon and is localized in the nucleus. - PMC - PubMed
- GenBank http://www.ncbi.nlm.nih.gov/Genbank/index.html DNA sequences of FGFs listed in Table 1 can be accessed through this website.
- HUGO Gene Nomenclature Database http://www.gene.ucl.ac.uk/nomenclature/ This database contains nomenclature and mapping information for human genes.
- Cytokine Family cDNA Database http://cytokine.medic.kumamoto-u.ac.jp/CFC/FGF/FGF.html This database contains nomenclature and mapping information for the FGF family.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases