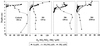Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes - PubMed (original) (raw)
Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes
G Braker et al. Appl Environ Microbiol. 2001 Apr.
Abstract
Steep vertical gradients of oxidants (O(2) and NO(3)(-)) in Puget Sound and Washington continental margin sediments indicate that aerobic respiration and denitrification occur within the top few millimeters to centimeters. To systematically explore the underlying communities of denitrifiers, Bacteria, and Archaea along redox gradients at distant geographic locations, nitrite reductase (nirS) genes and bacterial and archaeal 16S rRNA genes (rDNAs) were PCR amplified and analyzed by terminal restriction fragment length polymorphism (T-RFLP) analysis. The suitablility of T-RFLP analysis for investigating communities of nirS-containing denitrifiers was established by the correspondence of dominant terminal restriction fragments (T-RFs) of nirS to computer-simulated T-RFs of nirS clones. These clones belonged to clusters II, III, and IV from the same cores and were analyzed in a previous study (G. Braker, J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje, Appl. Environ. Microbiol. 66:2096-2104, 2000). T-RFLP analysis of nirS and bacterial rDNA revealed a high level of functional and phylogenetic diversity, whereas the level of diversity of Archaea was lower. A comparison of T-RFLPs based on the presence or absence of T-RFs and correspondence analysis based on the frequencies and heights of T-RFs allowed us to group sediment samples according to the sampling location and thus clearly distinguish Puget Sound and the Washington margin populations. However, changes in community structure within sediment core sections during the transition from aerobic to anaerobic conditions were minor. Thus, within the top layers of marine sediments, redox gradients seem to result from the differential metabolic activities of populations of similar communities, probably through mixing by marine invertebrates rather than from the development of distinct communities.
Figures
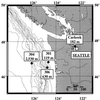
FIG. 1
Locations and water depths of the sampling stations at Puget Sound and the Washington margin.
FIG. 2
Profiles of the oxidants oxygen, nitrate, nitrite, and ammonium within sediment cores from Puget Sound (Carkeek) and the Washington margin (cores 301, 306, and 304).
FIG. 3
Total numbers of T-RFs derived from amplified nirS genes and 16S rDNAs (Bacteria and Archaea) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Total numbers were calculated from cleavages with three restriction endonucleases.
FIG. 4
Relative abundances of T-RF of amplified nirS genes (A) and 16S rDNAs of Bacteria (B) and Archaea (C) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Diagrams show results after cleavage with _Hha_I (nirS), _Hae_III (Bacteria), and _Hha_I (Archaea). Numbers on top of the columns indicate the numbers of detectable T-RFs for individual samples. Numbers in the keys indicate the lengths of the T-RFs in base pairs for fragments with a relative abundance of more than 2%.
FIG. 5
Relationship of T-RFLPs of amplified nirS genes and 16S rDNAs (Bacteria and Archaea) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Dendrograms calculated from the presence and absence of peaks (A) and correspondence analysis using combined results from three individual cleavages (B) are shown. Dim, dimension.
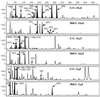
FIG. 6
Comparison of T-RFs of amplified nirS genes to nirS clone fragments from in silico digestion of sequences from marine sediment samples. T-RFs were detected in sediment samples from Puget Sound (C/1) and the Washington margin (304/1); cloned nirS fragments were obtained from environmental DNA extracts from the same core. Shaded peaks and arrows indicate clones corresponding to T-RFs. x axis, size (base pairs); y axis, relative fluorescence units.
Similar articles
- Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in pacific northwest marine sediment communities.
Braker G, Zhou J, Wu L, Devol AH, Tiedje JM. Braker G, et al. Appl Environ Microbiol. 2000 May;66(5):2096-104. doi: 10.1128/AEM.66.5.2096-2104.2000. Appl Environ Microbiol. 2000. PMID: 10788387 Free PMC article. - Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea.
Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. Swan BK, et al. Appl Environ Microbiol. 2010 Feb;76(3):757-68. doi: 10.1128/AEM.02409-09. Epub 2009 Nov 30. Appl Environ Microbiol. 2010. PMID: 19948847 Free PMC article. - [Phylogeny diversity of the nitrite reductase gene (nirS) in the sediments of the eutrophic East Lake, Wuhan].
Cheng Z, Yang J, Li H, Zhu B, Chen X, Yan Y. Cheng Z, et al. Wei Sheng Wu Xue Bao. 2011 May;51(5):667-75. Wei Sheng Wu Xue Bao. 2011. PMID: 21800630 Chinese. - Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific.
Castro-González M, Braker G, Farías L, Ulloa O. Castro-González M, et al. Environ Microbiol. 2005 Sep;7(9):1298-306. doi: 10.1111/j.1462-2920.2005.00809.x. Environ Microbiol. 2005. PMID: 16104853 - Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea.
Li M, Hong Y, Cao H, Gu JD. Li M, et al. Microb Ecol. 2013 Aug;66(2):281-96. doi: 10.1007/s00248-012-0175-y. Epub 2013 Jan 29. Microb Ecol. 2013. PMID: 23354291
Cited by
- Spatio-temporal shifts in community structure and activity of nirS-type denitrifiers in the sediment cores of Pearl River Estuary.
Xie H, Hong Y, Liu H, Jiao L, Wu J, Wang L. Xie H, et al. PLoS One. 2020 Apr 21;15(4):e0231271. doi: 10.1371/journal.pone.0231271. eCollection 2020. PLoS One. 2020. PMID: 32315323 Free PMC article. - DNA finger printing of S. Mutans present in the saliva of caries active children and those associated with intellectual disability - An RAPD analysis.
Prabhakar AR, Sreeja G, Naik SV. Prabhakar AR, et al. Saudi Dent J. 2019 Oct;31(4):424-430. doi: 10.1016/j.sdentj.2019.04.009. Epub 2019 Apr 27. Saudi Dent J. 2019. PMID: 31700219 Free PMC article. - Using laboratory-generated biosolids to evaluate the microbial ecotoxicity of triclosan in a simulated land application scenario.
Holzem RM, Gardner CM, Stapleton HM, Gunsch CK. Holzem RM, et al. Environ Sci Pollut Res Int. 2018 Apr;25(11):11084-11099. doi: 10.1007/s11356-017-1147-z. Epub 2018 Feb 6. Environ Sci Pollut Res Int. 2018. PMID: 29411281 - The effect of bio-irrigation by the polychaete Lanice conchilega on active denitrifiers: Distribution, diversity and composition of nosZ gene.
Yazdani Foshtomi M, Leliaert F, Derycke S, Willems A, Vincx M, Vanaverbeke J. Yazdani Foshtomi M, et al. PLoS One. 2018 Feb 6;13(2):e0192391. doi: 10.1371/journal.pone.0192391. eCollection 2018. PLoS One. 2018. PMID: 29408934 Free PMC article. - Capturing Compositional Variation in Denitrifying Communities: a Multiple-Primer Approach That Includes Epsilonproteobacteria.
Murdock SA, Juniper SK. Murdock SA, et al. Appl Environ Microbiol. 2017 Mar 2;83(6):e02753-16. doi: 10.1128/AEM.02753-16. Print 2017 Mar 15. Appl Environ Microbiol. 2017. PMID: 28087525 Free PMC article.
References
- Ausubel F M, Brent R, Kinston E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991.
- Brandes J A, Devol A H. Simultaneous nitrate and oxygen respiration in coastal sediments: evidence for discrete diagenesis. J Mar Res. 1995;52:771–797.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources