Lineage-specific gene expansions in bacterial and archaeal genomes - PubMed (original) (raw)
Comparative Study
Lineage-specific gene expansions in bacterial and archaeal genomes
I K Jordan et al. Genome Res. 2001 Apr.
Abstract
Gene duplication is an important mechanistic antecedent to the evolution of new genes and novel biochemical functions. In an attempt to assess the contribution of gene duplication to genome evolution in archaea and bacteria, clusters of related genes that appear to have expanded subsequent to the diversification of the major prokaryotic lineages (lineage-specific expansions) were analyzed. Analysis of 21 completely sequenced prokaryotic genomes shows that lineage-specific expansions comprise a substantial fraction (approximately 5%-33%) of their coding capacities. A positive correlation exists between the fraction of the genes taken up by lineage-specific expansions and the total number of genes in a genome. Consistent with the notion that lineage-specific expansions are made up of relatively recently duplicated genes, >90% of the detected clusters consists of only two to four genes. The more common smaller clusters tend to include genes with higher pairwise similarity (as reflected by average score density) than larger clusters. Regardless of size, cluster members tend to be located more closely on bacterial chromosomes than expected by chance, which could reflect a history of tandem gene duplication. In addition to the small clusters, almost all genomes also contain rare large clusters of size > or =20. Several examples of the potential adaptive significance of these large clusters are explored. The presence or absence of clusters and their related genes was used as the basis for the construction of a similarity graph for completely sequenced prokaryotic genomes. The topology of the resulting graph seems to reflect a combined effect of common ancestry, horizontal transfer, and lineage-specific gene loss.
Figures
Figure 1
Cluster sizes in number of genes (_Y_-axis, gray bars) for four representative species, (A) Campylobacter jejuni, (B) Methanococcus janaschii, (C) Mycoplasma pneumoniae, and (D) Treponema pallidum, compared to the average number of best hits (BeTs) for each cluster (_Y_-axis, black bars) in all other completely sequenced bacterial genomes.
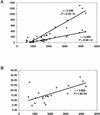
Figure 2
Linear correlation between genome size (in number of genes) and the parameters of lineage-specific expansions. Correlation coefficients (r) and significance levels (P) were determined using ordinary least squares linear regression. (A) For completely sequenced prokaryotic genomes, genome size (_X_-axis) is plotted against the number of genes in lineage-specific clusters (diamonds) and the number of such clusters (squares). (B) Genome size (_X_-axis) is plotted against the percentage of the genome made up of lineage-specific clusters (triangles).
Figure 3
Frequency distribution (99% quantile) of lineage-specific expansion cluster sizes (_X_-axis in numbers of genes). Observed data are shown with diamonds. These data were fit using the logarithmic approximation (line).
Figure 4
Linear correlation between cluster size in number of genes (_X_-axis) and average score density per cluster (_Y_-axis). Correlation coefficients (r) and significance levels (P) were determined using ordinary least squares linear regression. Removal of the two largest clusters (size 67 and 90) results in a greater magnitude of r and a lower P value (i.e., a stronger negative correlation).
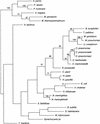
Figure 5
Maximum parsimony graph for completely sequenced archaeal and bacterial genomes. The root was provisionally placed between archaea and bacteria.
Similar articles
- Genome trees constructed using five different approaches suggest new major bacterial clades.
Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. Wolf YI, et al. BMC Evol Biol. 2001 Oct 20;1:8. doi: 10.1186/1471-2148-1-8. BMC Evol Biol. 2001. PMID: 11734060 Free PMC article. - Origin and evolution of gene families in Bacteria and Archaea.
Collins RE, Merz H, Higgs PG. Collins RE, et al. BMC Bioinformatics. 2011 Oct 5;12 Suppl 9(Suppl 9):S14. doi: 10.1186/1471-2105-12-S9-S14. BMC Bioinformatics. 2011. PMID: 22151831 Free PMC article. - Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context.
Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Wolf YI, et al. Genome Res. 2001 Mar;11(3):356-72. doi: 10.1101/gr.gr-1619r. Genome Res. 2001. PMID: 11230160 - Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.
Koonin EV, Wolf YI. Koonin EV, et al. Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review. - Selection for gene clustering by tandem duplication.
Reams AB, Neidle EL. Reams AB, et al. Annu Rev Microbiol. 2004;58:119-42. doi: 10.1146/annurev.micro.58.030603.123806. Annu Rev Microbiol. 2004. PMID: 15487932 Review.
Cited by
- The origin of eukaryotes is suggested as the symbiosis of pyrococcus into gamma-proteobacteria by phylogenetic tree based on gene content.
Horiike T, Hamada K, Miyata D, Shinozawa T. Horiike T, et al. J Mol Evol. 2004 Nov;59(5):606-19. doi: 10.1007/s00239-004-2652-5. J Mol Evol. 2004. PMID: 15693617 - Proteome and allergenome of the European house dust mite Dermatophagoides pteronyssinus.
Waldron R, McGowan J, Gordon N, McCarthy C, Mitchell EB, Fitzpatrick DA. Waldron R, et al. PLoS One. 2019 May 1;14(5):e0216171. doi: 10.1371/journal.pone.0216171. eCollection 2019. PLoS One. 2019. PMID: 31042761 Free PMC article. - Gene Expansion and Positive Selection as Bacterial Adaptations to Oligotrophic Conditions.
Props R, Monsieurs P, Vandamme P, Leys N, Denef VJ, Boon N. Props R, et al. mSphere. 2019 Feb 6;4(1):e00011-19. doi: 10.1128/mSphereDirect.00011-19. mSphere. 2019. PMID: 30728279 Free PMC article. - The SNAP hypothesis: Chromosomal rearrangements could emerge from positive Selection during Niche Adaptation.
Brandis G, Hughes D. Brandis G, et al. PLoS Genet. 2020 Mar 4;16(3):e1008615. doi: 10.1371/journal.pgen.1008615. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32130223 Free PMC article. - Genome-Wide Identification of Calcium Dependent Protein Kinase Gene Family in Plant Lineage Shows Presence of Novel D-x-D and D-E-L Motifs in EF-Hand Domain.
Mohanta TK, Mohanta N, Mohanta YK, Bae H. Mohanta TK, et al. Front Plant Sci. 2015 Dec 24;6:1146. doi: 10.3389/fpls.2015.01146. eCollection 2015. Front Plant Sci. 2015. PMID: 26734045 Free PMC article.
References
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. - PubMed
- Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. - PubMed
- Bailey TL, Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics. 1998;14:48–54. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources