Automated finishing with autofinish - PubMed (original) (raw)
Comparative Study
Automated finishing with autofinish
D Gordon et al. Genome Res. 2001 Apr.
Abstract
Currently, the genome sequencing community is producing shotgun sequence data at a very high rate, but finishing (collecting additional directed sequence data to close gaps and improve the quality of the data) is not matching that rate. One reason for the difference is that shotgun sequencing is highly automated but finishing is not: Most finishing decisions, such as which directed reads to obtain and which specialized sequencing techniques to use, are made by people. If finishing rates are to increase to match shotgun sequencing rates, most finishing decisions also must be automated. The Autofinish computer program (which is part of the computer software package) does this by automatically choosing finishing reads. Autofinish is able to suggest most finishing reads required for completion of each sequencing project, greatly reducing the amount of human attention needed. sometimes completely finishes the project, with no human decisions required. It cannot solve the most complex problems, so we recommend that Autofinish be allowed to suggest reads for the first three rounds of finishing, and if the project still is not finished completely, a human finisher complete the work. We compared this Autofinish-Hybrid method of finishing against a human finisher in five different projects with a variety of shotgun depths by finishing each project twice--once with each method. This comparison shows that the Autofinish-Hybrid method saves many hours over a human finisher alone, while using roughly the same number and type of reads and closing gaps at roughly the same rate. Autofinish currently is in production use at several large sequencing centers. It is designed to be adaptable to the finishing strategy of the lab--it can finish using some or all of the following: resequencing reads, reverses, custom primer walks on either subclone templates or whole clone templates, PCR, or minilibraries. Autofinish has been used for finishing cDNA, genomic clones, and whole bacterial genomes (see http://www.phrap.org).
Figures
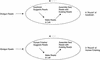
Figure 1
Finishing procedures with
Autofinish
and with a human finisher.
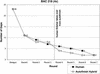
Figure 2
Gap Closing:
Autofinish
-Hybrid vs. Human-only for five different BACs.
Figure 2
Gap Closing:
Autofinish
-Hybrid vs. Human-only for five different BACs.
Figure 2
Gap Closing:
Autofinish
-Hybrid vs. Human-only for five different BACs.
Figure 2
Gap Closing:
Autofinish
-Hybrid vs. Human-only for five different BACs.
Figure 2
Gap Closing:
Autofinish
-Hybrid vs. Human-only for five different BACs.
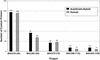
Figure 3
Finishing reads required:
Autofinish
-Hybrid vs. Human-only.
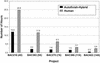
Figure 4
PCR reads required:
Autofinish
-Hybrid vs. Human-only.
Figure 5
Custom primers required:
Autofinish
-Hybrid vs. Human-only.
Figure 6
Human hours required for choosing reads:
Autofinish
-Hybrid vs. Human-only.
Figure 7
Autofinish
checks each contig to see if either end is the clone end. Masked clone vector and subclone vector both appear as Xs, and Bs are high quality bases that do not match subclone or clone vector. Notice that vector-insert junctions of reads 1–4 are aligned. If read 5 were not present, this figure would suggest a typical clone end with the Xs clone vector. If only read 1 and read 2 were present, the Xs could instead be subclone vector, which just happens to align; but the presence of two additional reads (read 3 and read 4), or additional vector bases in one read, would make this less likely. However, if read 5 were present (all high quality bases surrounding the putative vector/insert junction), it would be unlikely that this is the clone end.
Similar articles
- Whole genome shotgun sequencing guided by bioinformatics pipelines--an optimized approach for an established technique.
Kaiser O, Bartels D, Bekel T, Goesmann A, Kespohl S, Pühler A, Meyer F. Kaiser O, et al. J Biotechnol. 2003 Dec 19;106(2-3):121-33. doi: 10.1016/j.jbiotec.2003.08.008. J Biotechnol. 2003. PMID: 14651855 - Consed: a graphical tool for sequence finishing.
Gordon D, Abajian C, Green P. Gordon D, et al. Genome Res. 1998 Mar;8(3):195-202. doi: 10.1101/gr.8.3.195. Genome Res. 1998. PMID: 9521923 - A novel approach to sequence validating protein expression clones with automated decision making.
Taycher E, Rolfs A, Hu Y, Zuo D, Mohr SE, Williamson J, Labaer J. Taycher E, et al. BMC Bioinformatics. 2007 Jun 13;8:198. doi: 10.1186/1471-2105-8-198. BMC Bioinformatics. 2007. PMID: 17567908 Free PMC article. - One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly.
Koren S, Phillippy AM. Koren S, et al. Curr Opin Microbiol. 2015 Feb;23:110-20. doi: 10.1016/j.mib.2014.11.014. Epub 2014 Dec 1. Curr Opin Microbiol. 2015. PMID: 25461581 Review. - Sequence assembly using next generation sequencing data--challenges and solutions.
Chin FY, Leung HC, Yiu SM. Chin FY, et al. Sci China Life Sci. 2014 Nov;57(11):1140-8. doi: 10.1007/s11427-014-4752-9. Epub 2014 Oct 17. Sci China Life Sci. 2014. PMID: 25326069 Review.
Cited by
- Horizontal gene transfer in Histophilus somni and its role in the evolution of pathogenic strain 2336, as determined by comparative genomic analyses.
Siddaramappa S, Challacombe JF, Duncan AJ, Gillaspy AF, Carson M, Gipson J, Orvis J, Zaitshik J, Barnes G, Bruce D, Chertkov O, Detter JC, Han CS, Tapia R, Thompson LS, Dyer DW, Inzana TJ. Siddaramappa S, et al. BMC Genomics. 2011 Nov 23;12:570. doi: 10.1186/1471-2164-12-570. BMC Genomics. 2011. PMID: 22111657 Free PMC article. - Clusters of adaptive evolution in the human genome.
Scheinfeldt LB, Biswas S, Madeoy J, Connelly CF, Akey JM. Scheinfeldt LB, et al. Front Genet. 2011 Sep 9;2:50. doi: 10.3389/fgene.2011.00050. eCollection 2011. Front Genet. 2011. PMID: 22303346 Free PMC article. - Nucleotide diversity and linkage disequilibrium in cold-hardiness- and wood quality-related candidate genes in Douglas fir.
Krutovsky KV, Neale DB. Krutovsky KV, et al. Genetics. 2005 Dec;171(4):2029-41. doi: 10.1534/genetics.105.044420. Epub 2005 Sep 12. Genetics. 2005. PMID: 16157674 Free PMC article. - Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps.
Tsai IJ, Otto TD, Berriman M. Tsai IJ, et al. Genome Biol. 2010;11(4):R41. doi: 10.1186/gb-2010-11-4-r41. Epub 2010 Apr 13. Genome Biol. 2010. PMID: 20388197 Free PMC article. - Genome reduction in Leptospira borgpetersenii reflects limited transmission potential.
Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. Bulach DM, et al. Proc Natl Acad Sci U S A. 2006 Sep 26;103(39):14560-5. doi: 10.1073/pnas.0603979103. Epub 2006 Sep 14. Proc Natl Acad Sci U S A. 2006. PMID: 16973745 Free PMC article.
References
- Ewing B, Hillier L, Wendl M, Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. - PubMed
- Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. - PubMed
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous