A comprehensive two-hybrid analysis to explore the yeast protein interactome - PubMed (original) (raw)
A comprehensive two-hybrid analysis to explore the yeast protein interactome
T Ito et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Protein-protein interactions play crucial roles in the execution of various biological functions. Accordingly, their comprehensive description would contribute considerably to the functional interpretation of fully sequenced genomes, which are flooded with novel genes of unpredictable functions. We previously developed a system to examine two-hybrid interactions in all possible combinations between the approximately 6,000 proteins of the budding yeast Saccharomyces cerevisiae. Here we have completed the comprehensive analysis using this system to identify 4,549 two-hybrid interactions among 3,278 proteins. Unexpectedly, these data do not largely overlap with those obtained by the other project [Uetz, P., et al. (2000) Nature (London) 403, 623-627] and hence have substantially expanded our knowledge on the protein interaction space or interactome of the yeast. Cumulative connection of these binary interactions generates a single huge network linking the vast majority of the proteins. Bioinformatics-aided selection of biologically relevant interactions highlights various intriguing subnetworks. They include, for instance, the one that had successfully foreseen the involvement of a novel protein in spindle pole body function as well as the one that may uncover a hitherto unidentified multiprotein complex potentially participating in the process of vesicular transport. Our data would thus significantly expand and improve the protein interaction map for the exploration of genome functions that eventually leads to thorough understanding of the cell as a molecular system.
Figures
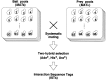
Figure 1
Outline of the comprehensive two-hybrid analysis. We cloned almost all yeast ORFs individually as a DNA-binding domain fusion (bait) in a_MAT_a strain and as an activation domain fusion (prey) in a MAT_α strain, and subsequently divided them into pools, each containing 96 clones. These bait and prey clone pools were systematically mated with each other, and the diploid cells formed were selected for the simultaneous activation of three reporter genes (ADE2, HIS3, and_URA3) followed by sequence tagging to obtain ISTs.
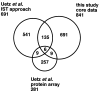
Figure 2
Overlap among the results of large-scale two-hybrid projects. The Venn diagram indicates the overlap among the core data of our analysis and those obtained by the high-throughput IST analysis and the protein array approach by Uetz et al. (11).
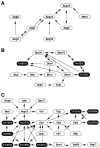
Figure 3
Biologically intriguing interaction networks. Three subnetworks consisting of proteins involved in autophagy (A), spindle pole body function (B), and vesicular transport (C) are shown. The arrows indicate the orientation of each two-hybrid interaction, beginning from the bait to the prey. Hypothetical proteins of unknown functions are indicated by black ovals with white letters.
Comment in
- Networking proteins in yeast.
Hazbun TR, Fields S. Hazbun TR, et al. Proc Natl Acad Sci U S A. 2001 Apr 10;98(8):4277-8. doi: 10.1073/pnas.091096398. Proc Natl Acad Sci U S A. 2001. PMID: 11296274 Free PMC article. No abstract available.
Similar articles
- Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins.
Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Ito T, et al. Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1143-7. doi: 10.1073/pnas.97.3.1143. Proc Natl Acad Sci U S A. 2000. PMID: 10655498 Free PMC article. - Roles for the two-hybrid system in exploration of the yeast protein interactome.
Ito T, Ota K, Kubota H, Yamaguchi Y, Chiba T, Sakuraba K, Yoshida M. Ito T, et al. Mol Cell Proteomics. 2002 Aug;1(8):561-6. doi: 10.1074/mcp.r200005-mcp200. Mol Cell Proteomics. 2002. PMID: 12376571 Review. - Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens.
Fromont-Racine M, Rain JC, Legrain P. Fromont-Racine M, et al. Nat Genet. 1997 Jul;16(3):277-82. doi: 10.1038/ng0797-277. Nat Genet. 1997. PMID: 9207794 - Building a protein interaction map: research in the post-genome era.
Chen Z, Han M. Chen Z, et al. Bioessays. 2000 Jun;22(6):503-6. doi: 10.1002/(SICI)1521-1878(200006)22:6<503::AID-BIES2>3.0.CO;2-7. Bioessays. 2000. PMID: 10842303 Review. - A network of protein-protein interactions in yeast.
Schwikowski B, Uetz P, Fields S. Schwikowski B, et al. Nat Biotechnol. 2000 Dec;18(12):1257-61. doi: 10.1038/82360. Nat Biotechnol. 2000. PMID: 11101803
Cited by
- Binding interface prediction by combining protein-protein docking results.
Hwang H, Vreven T, Weng Z. Hwang H, et al. Proteins. 2014 Jan;82(1):57-66. doi: 10.1002/prot.24354. Epub 2013 Aug 31. Proteins. 2014. PMID: 23836482 Free PMC article. - Pre-trained protein language model sheds new light on the prediction of Arabidopsis protein-protein interactions.
Zhou K, Lei C, Zheng J, Huang Y, Zhang Z. Zhou K, et al. Plant Methods. 2023 Dec 7;19(1):141. doi: 10.1186/s13007-023-01119-6. Plant Methods. 2023. PMID: 38062445 Free PMC article. - Drosophila protein interaction map (DPiM): a paradigm for metazoan protein complex interactions.
Guruharsha KG, Obar RA, Mintseris J, Aishwarya K, Krishnan RT, Vijayraghavan K, Artavanis-Tsakonas S. Guruharsha KG, et al. Fly (Austin). 2012 Oct-Dec;6(4):246-53. doi: 10.4161/fly.22108. Fly (Austin). 2012. PMID: 23222005 Free PMC article. - Integrative approaches for predicting protein function and prioritizing genes for complex phenotypes using protein interaction networks.
Ma X, Chen T, Sun F. Ma X, et al. Brief Bioinform. 2014 Sep;15(5):685-98. doi: 10.1093/bib/bbt041. Epub 2013 Jun 19. Brief Bioinform. 2014. PMID: 23788799 Free PMC article. - Cell-free Determination of Binary Complexes That Comprise Extended Protein-Protein Interaction Networks of Yersinia pestis.
Keasey SL, Natesan M, Pugh C, Kamata T, Wuchty S, Ulrich RG. Keasey SL, et al. Mol Cell Proteomics. 2016 Oct;15(10):3220-3232. doi: 10.1074/mcp.M116.059337. Epub 2016 Aug 3. Mol Cell Proteomics. 2016. PMID: 27489291 Free PMC article.
References
- Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. - PubMed
- Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:563–567. - PubMed
- Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. - PubMed
- Ogasawara N. Res Microbiol. 2000;151:129–134. - PubMed
- Fraser A G, Kamath R S, Zipperien P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases