Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1 - PubMed (original) (raw)
Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1
K Narayanan et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Cells of the craniofacial skeleton are derived from a common mesenchymal progenitor. The regulatory factors that control their differentiation into various cell lineages are unknown. To investigate the biological function of dentin matrix protein 1 (DMP1), an extracellular matrix gene involved in calcified tissue formation, stable transgenic cell lines and adenovirally infected cells overexpressing DMP1 were generated. The findings in this paper demonstrate that overexpression of DMP1 in pluripotent and mesenchyme-derived cells such as C3H10T1/2, MC3T3-E1, and RPC-C2A can induce these cells to differentiate and form functional odontoblast-like cells. Functional differentiation of odontoblasts requires unique sets of genes being turned on and off in a growth- and differentiation-specific manner. The genes studied include transcription factors like core binding factor 1 (Cbfa1), bone morphogenetic protein 2 (BMP2), and BMP4; early markers for extracellular matrix deposition like alkaline phosphatase (ALP), osteopontin, osteonectin, and osteocalcin; and late markers like DMP2 and dentin sialoprotein (DSP) that are expressed by terminally differentiated odontoblasts and are responsible for the formation of tissue-specific dentin matrix. However, this differentiation pathway was limited to mesenchyme-derived cells only. Other cell lines tested by the adenoviral expression system failed to express odontoblast-phenotypic specific genes. An in vitro mineralized nodule formation assay demonstrated that overexpressed cells could differentiate and form a mineralized matrix. Furthermore, we also demonstrate that phosphorylation of Cbfa1 (osteoblast-specific transcription factor) was not required for the expression of odontoblast-specific genes, indicating the involvement of other unidentified odontoblast-specific transcription factors or coactivators. Cell lines that differentiate into odontoblast-like cells are useful tools for studying the mechanism involved in the terminal differentiation process of these postmitotic cells.
Figures
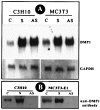
Figure 1
Overexpression of DMP1. The cell lines were transfected as described in_Materials and Methods_. Stable selection was made at 800 μg/ml of G418 sulfate. The expression level of DMP1 was analyzed by either Northern blot (A) or Western blot (B). Twenty micrograms of total RNA was resolved on a 0.8% agarose gel under denaturing conditions and transferred. The membrane was probed with random [32P]dCTP-labeled DMP1 cDNA or glyceraldehyde-3-phosphate dehydrogenase. Fifteen micrograms of total protein was resolved on a 10% SDS-PAGE gel and subsequently electroblotted. The membrane was probed with affinity-purified anti-DMP1 antibody (B).
Figure 2
DMP1 overexpression and morphology. The G418-resistant cells were plated at high density (3 × 106 cells per milliliter) and low density (1 × 104 cells per milliliter). The cells were allowed to grow in the appropriate medium for 24 h, and photographs were taken with a phase-contrast microscope. “Mock” represents a stable cell line expressing the “empty” vector (A). “Sense” (B) and “antisense” (C) represent the stable cell lines overexpressing sense and antisense DMP1 cDNA. Microphotographs were taken at ×10 with a Nikon camera.
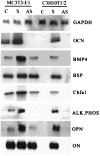
Figure 3
Northern analysis of various marker genes. Total RNA was isolated from G418-resistant cell lines overexpressing DMP1. Northern blot was carried out with 20 μg of RNA. The probes were randomly labeled with [32P]dCTP. C, Stable MC3T3-E1 and C3H10T1/2 cell lines expressing the “empty vector.” S, Stable MC3T3-E1 and C3H10T1/2 cell lines overexpressing DMP1 in sense orientation. AS, Stable MC3T3-E1 and C3H10T1/2 cell lines overexpressing DMP1 in the antisense orientation. Glyceraldehyde-3-phosphate dehydrogenase was used as the control.
Figure 4
Expression of DSP and DMP2. G418-resistant cells were analyzed by RT-PCR as described in Materials and Methods. C, Mock MC3T3-E1 and C3H10T1/2. S, Stable MC3T3-E1 and C3H10T1/2 cell lines overexpressing DMP1 (sense). AS, Cell lines overexpressing the antisense sequence of DMP1. PCR products were resolved on a 2% agarose gel.
Figure 5
Suppression of DSP and DMP2 by antisense DMP1. Cell lines overexpressing DMP1 (sense) were infected with Ad5/CMV-DMP1 (antisense) as described in Materials and Methods. Total RNA was isolated (after 24 h), and RT-PCR was performed with gene-specific primers. C, Control cells (with “empty vector”). S, cells overexpressing sense DMP1. S + AS infected with Ad5-CMV-DMP1 (antisense) on cells overexpressing DMP1 (sense). In each case DSP and DMP2 were analyzed by RT-PCR.
Figure 6
Cell specificity. To study cell specificity, the Ad5/CMV-DMP1 (sense) and Ad5/CMV-lacZ adenoviruses were used. Different cell lines were infected with adenovirus for 3 h, and fresh medium was then added. Total RNA was isolated (after 24 h), and RT-PCR performed as described in Materials and Methods. C, Control cells infected with Ad5/CMV-lacZ. +, Cells infected with Ad5/CMV-DMP1 (sense). Hypoxanthine phosphoribosyltransferase was used as the control, and gene expression levels of Cbfa1, OCN, DSP, DMP2, osteonectin, and OPN were analyzed.
Figure 7
Signaling pathway in DSP/DMP2 expression. (A) DMP1 phosphorylation. C3H10T1/2 cells were infected with adenoviral constructs. Total protein extracts were prepared as described in_Materials and Methods_ and immunoprecipitated with affinity-purified anti-DMP1 antibody. (Top) Western blot against anti-phosphoserine antibody on DMP1 immunoprecipitates. (Bottom) Western blot performed with anti-DMP1 antibody on the DMP1 immunoprecipitates. (B) DSP and DMP2 expression. Cells (2 × 106 cells per milliliter) were seeded 24 h before infection with Ad5/CMV-DMP1. Infection was carried out for 3 h, and fresh medium was added to the cells. A MAPK inhibitor (PD98059, 100 μg/ml) and a casein kinase II inhibitor (heparan sulfate, 100 μg/ml) were added along with fresh medium and incubated for another 24 h. Total RNA was isolated from cells, and RT-PCR was performed. C, Control cells infected with β-gal expression cassette. +, Positive control used for the experiment [infected with Ad5/CMV-DMP1 (sense) without inhibitors]. 1 and 2, Duplicates for the experiment. RT-PCR analysis for Cbfa1, OCN, DSP, and DMP2 was done with hypoxanthine phosphoribosyltransferase as a control.
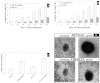
Figure 8
DMP1 overexpression and mineralization: a graphical representation of the effect of DMP1 overexpression on mineralized nodule formation in MC3T3-E1 (A) and C3H10T1/2 cells (B). (C) The size of the mineralized nodule was measured with an occulometer. Mock cells expressing the empty vector were used as a control. (D) The mineralized nodule was microphotographed (after 15 days) with a Nikon camera at ×10.
Similar articles
- Molecular insights into the lineage-specific determination of odontoblasts: the role of Cbfa1.
Gaikwad JS, Hoffmann M, Cavender A, Bronckers AL, D'Souza RN. Gaikwad JS, et al. Adv Dent Res. 2001 Aug;15:19-24. doi: 10.1177/08959374010150010501. Adv Dent Res. 2001. PMID: 12640733 - Differentiation of Odontoblast-Like Cells From Mouse Induced Pluripotent Stem Cells by Pax9 and Bmp4 Transfection.
Seki D, Takeshita N, Oyanagi T, Sasaki S, Takano I, Hasegawa M, Takano-Yamamoto T. Seki D, et al. Stem Cells Transl Med. 2015 Sep;4(9):993-7. doi: 10.5966/sctm.2014-0292. Epub 2015 Jul 1. Stem Cells Transl Med. 2015. PMID: 26136503 Free PMC article. - Klf10 regulates odontoblast differentiation and mineralization via promoting expression of dentin matrix protein 1 and dentin sialophosphoprotein genes.
Chen Z, Li W, Wang H, Wan C, Luo D, Deng S, Chen H, Chen S. Chen Z, et al. Cell Tissue Res. 2016 Feb;363(2):385-98. doi: 10.1007/s00441-015-2260-2. Epub 2015 Aug 28. Cell Tissue Res. 2016. PMID: 26310138 Free PMC article. - The nature and functional significance of dentin extracellular matrix proteins.
Butler WT, Ritchie H. Butler WT, et al. Int J Dev Biol. 1995 Feb;39(1):169-79. Int J Dev Biol. 1995. PMID: 7626404 Review. - A Perspective on Stem Cells as Biological Systems that Produce Differentiated Osteoblasts and Odontoblasts.
Ariffin SH, Manogaran T, Abidin IZ, Wahab RM, Senafi S. Ariffin SH, et al. Curr Stem Cell Res Ther. 2017;12(3):247-259. doi: 10.2174/1574888X11666161026145149. Curr Stem Cell Res Ther. 2017. PMID: 27784228 Review.
Cited by
- Dentin matrix protein 1 and HUVEC-ECM scaffold promote the differentiation of human dental pulp stem cells into endothelial lineage: implications in regenerative medicine.
Ganapathy A, Narayanan K, Chen Y, Villani C, George A. Ganapathy A, et al. Front Physiol. 2024 Jul 8;15:1429247. doi: 10.3389/fphys.2024.1429247. eCollection 2024. Front Physiol. 2024. PMID: 39040080 Free PMC article. - Notum regulates the cusp and root patterns in mouse molar.
Adasooriya D, Jeong JK, Kyeong M, Kan S, Kim J, Cho ES, Cho SW. Adasooriya D, et al. Sci Rep. 2024 Jun 13;14(1):13633. doi: 10.1038/s41598-024-64340-w. Sci Rep. 2024. PMID: 38871845 Free PMC article. - Atp6i deficient mouse model uncovers transforming growth factor-β1 /Smad2/3 as a key signaling pathway regulating odontoblast differentiation and tooth root formation.
Wang J, McVicar A, Chen Y, Deng HW, Zhao Z, Chen W, Li YP. Wang J, et al. Int J Oral Sci. 2023 Aug 21;15(1):35. doi: 10.1038/s41368-023-00235-2. Int J Oral Sci. 2023. PMID: 37599332 Free PMC article. - Autologous Tooth Graft: Innovative Biomaterial for Bone Regeneration. Tooth Transformer® and the Role of Microbiota in Regenerative Dentistry. A Systematic Review.
Inchingolo AM, Patano A, Di Pede C, Inchingolo AD, Palmieri G, de Ruvo E, Campanelli M, Buongiorno S, Carpentiere V, Piras F, Settanni V, Viapiano F, Hazballa D, Rapone B, Mancini A, Di Venere D, Inchingolo F, Fatone MC, Palermo A, Minetti E, Lorusso F, Scarano A, Sauro S, Tartaglia GM, Bordea IR, Dipalma G, Malcangi G. Inchingolo AM, et al. J Funct Biomater. 2023 Feb 27;14(3):132. doi: 10.3390/jfb14030132. J Funct Biomater. 2023. PMID: 36976056 Free PMC article. Review. - Effects of Sr2+, BO33-, and SiO32- on Differentiation of Human Dental Pulp Stem Cells into Odontoblast-Like Cells.
Miyano Y, Mikami M, Katsuragi H, Shinkai K. Miyano Y, et al. Biol Trace Elem Res. 2023 Dec;201(12):5585-5600. doi: 10.1007/s12011-023-03625-z. Epub 2023 Mar 14. Biol Trace Elem Res. 2023. PMID: 36917393
References
- Erickson C A, Reedy M V. Curr Top Dev Biol. 1998;40:177–209. - PubMed
- Linde A, Goldberg M. Crit Rev Oral Biol Med. 1993;4:679–728. - PubMed
- Veis A. In: Extracellular Matrix. Comper W D, editor. London: Harwood; 1996. pp. 41–76.
- Dimuzio M T, Veis A. Calcif Tissue Res. 1978;25:169–178. - PubMed
- George A, Bannon L, Sabsay B, Dillon J W, Malone J, Veis A, Jenkins A N, Gilbert D J, Copeland N G. J Biol Chem. 1996;271:32869–32873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous