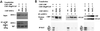Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity - PubMed (original) (raw)
Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity
E Soutoglou et al. EMBO J. 2001.
Abstract
CREB-binding protein (CBP) and CBP-associated factor (P/CAF) are coactivators possessing an intrinsic histone acetyltransferase (HAT) activity. They are positioned at promoter regions via association with sequence-specific DNA-binding factors and stimulate transcription in a gene-specific manner. The current view suggests that coactivator function depends mainly on the strength and specificity of transcription factor-coactivator interactions. Here we show that two dominant-negative mutants of hepatocyte nuclear factor-1alpha (HNF-1alpha), P447L and P519L, occurring in maturity onset diabetes of the young (MODY3) patients, exhibit paradoxically stronger interactions than the wild-type protein with either CBP or P/CAF. However, CBP and P/CAF recruited by these mutants lack HAT activity. In contrast, wild-type HNF-1alpha and other transcription factors, such as Sp1 or HNF-4, stimulated the HAT activity of CBP. The results suggest a more dynamic role for DNA-binding proteins in the transcription process than was considered previously. They are not only required for the recruitment of coactivators to the promoter but they may also modulate their enzymatic activity.
Figures
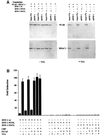
Fig. 1. Functional analysis of CBP and P/CAF interactions with HNF-1α mutants. (A) NIH 3T3 cells with stably integrated 3×AlbPE AdML-CAT were transfected with 0.5 µg of the indicated CMV-HNF-1α mutant plasmids, together with 2 µg of CMV-CBP or CMV-P/CAF expression vectors. The bars represent normalized CAT activities and standard errors from four independent experiments, expressed as fold induction above basal levels. Note the different scale of the ordinates. (B and C) Extracts from Cos-1 cells transfected with the indicated plasmids were immunoprecipitated (IP) with α-CBP or α-Flag antibodies, and the presence of HNF-1α in the immunoprecipitates was assessed in western blots by α-HNF-1α antibody. Input lanes represent 15% of the extracts used for immunoprecipitations.
Fig. 2. HNF-1α C-term P447L and P519L Gal4 fusion proteins recapitulate HNF-1α P447L and P519L full-length-mutant properties. (A) C33 cells were co-transfected with 1 µg of 5×Gal4 E1b-luc and 0.2 µg of Gal4-wt, Gal4-P447L or Gal4-P519L mutant expression vectors as indicated. (B) C33 cells were co-transfected with 1 µg of 5×Gal4 E1b-luc and 0.2 µg of Gal4-wt together with increasing amounts (0.4, 0.8 and 1.6 µg) of either Gal4, or Gal4 P447L or Gal4-P519L expression vectors as indicated. Bars represent normalized luciferase activity and standard errors from at least four independent experiments. The data are expressed as fold activation above basal levels. (C) In vitro pull-down experiments were performed with 35S-labeled, in vitro translated CBP and P/CAF probes. Full-length HNF-1α and its mutant derivatives were expressed as His6-tagged proteins and immobilized on Talon resin. As a control (Contr.), a column containing the unrelated HrpE protein was used. The C-terminal parts of HNF-1α and of its mutant derivatives were expressed as GST fusion proteins. Input lanes represent 15% of the probe used for the interactions. (D) 35S-labeled, in vitro translated full-length HNF-1α and its mutant derivatives were digested with the indicated amounts of V8 protease and analyzed by SDS–PAGE. Arrows indicate protease-induced bands detectable only with the mutant proteins.
Fig. 3. In vivo association of NCoR and HDAC-1 with HNF-1α mutants. (A) Cos-1 cells were transfected with the indicated plasmids and incubated with or without 1 µM TSA for 12 h before harvest. Nuclear extracts were prepared and immunoprecipitated with α-HNF-1α antibody. The precipitated proteins were analyzed in western blots using α-NCoR or α-HDAC-1 antibodies. (B) The effect of TSA treatment on HNF-1-mediated transcription. NIH-3T3 cells with stably integrated 3×AlbPE AdML-CAT were transfected by 0.5 µg of wild-type CMV-HNF-1α, CMV-HNF-1α P519L or CMV-HNF-1α P447L, together with 2 µg of CMV-CBP, CMV-P/CAF or CMV-CBP + CMVP/CAF expression vectors. Where indicated, the cells were treated with 1 µM TSA for 12 h, before harvesting. The bars represent normalized CAT activities and standard errors from three independent experiments, expressed as fold inductions above basal levels.
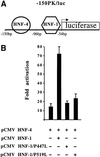
Fig. 4. _Trans_-acting effect of HNF-1α P447L and P519L mutants on the transcriptional activity of the L-PK gene promoter. (A) Schematic representation of the –150PK/luc construct corresponding to the proximal regulatory regions of the L-PK gene promoter. The –150PK/luc construct consists of HNF-1- and HNF-4-binding sites in front of the luciferase reporter gene. (B) C33 cells were co-transfected with 1 µg of –150PK/luc, 1 µg of CMV HNF-4 and 1 µg of CMV HNF-1, CMV HNF-1 P447L or CMV HNF-1 P519L expression vectors as indicated. Bars represent normalized luciferase activity and standard error from at least three independent experiments. The data are expressed as fold activation above basal level. Statistical significance was determined by a two-tailed Student’s _t_-test.
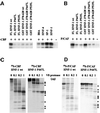
Fig. 5. Analysis of the in vivo formation of the HNF-1α–CBP–P/CAF trimeric complex and the HAT activities of CBP and P/CAF associated with HNF-1α proteins. (A) Extracts from Cos-1 cells transfected with the indicated plasmids were first immunoprecipitated with α-CBP antibody. The immune complexes were eluted from the protein A–Sepharose column by incubating the beads with 0.1 mg/ml CBP peptide (Santa Cruz Biotechnologies). The eluates were subjected to a second immunoprecipitation with α-Flag antibody. HNF-1α proteins in the HNF-1α–CBP–P/CAF trimeric complex form were detected in the resulting precipitates by western blot analysis using α-HNF-1α antibody. The input panel represents 5% of the amounts of extracts used for the first immunoprecipitation. (B) Extracts from Cos-1 cells transfected by the indicated plasmids were immunoprecipitated with α-HNF-1α antibody. The amounts of CBP and P/CAF proteins in the immunoprecipitates were estimated by western blot analysis (upper panels), and their corresponding HAT activities were assessed by IP-HAT assay (lower panel).
Fig. 6. Analysis of the effects of HNF-1α and its mutant derivatives on the HAT activity and conformation of CBP and P/CAF, in vitro. A 60 ng aliquot of baculovirus-expressed CBP (A) or P/CAF (B) was incubated with 1 µg of the indicated recombinant proteins in 1× HAT buffer, for 1 h at 4°C. After the addition of core histones and [3H]acetyl-CoA, the samples were incubated further at 30°C for 1 h and analyzed by SDS–PAGE. (C and D) CBP and P/CAF were translated in vitro in a 10 times scaled-up reaction and allowed to interact with Talon resin-bound full-length wild-type HNF-1α or P447L mutant. After extensive washing, proteins remaining in the beads were digested with the indicated amounts of V8 protease. Dots on the right indicate protease-induced bands.
Fig. 7. Analysis of the HNF-1α-dependent recruitment of CBP and P/CAF to chromatin template in intact cells and the acetylation state of the neighboring nucleosomes. Soluble chromatin from formaldehyde-fixed NIH 3T3 cells transfected with the indicated plasmids was prepared and immunoprecipitated with (A) α-HNF-1α, α-CBP or α-P/CAF, (B) anti-acetyl H3 or anti-acetyl H4 antibodies. The radioactivities of the 271 bp PCR products were quantitated by phosphoimage analysis. The data are expressed as percentages of the normalized radioactivities obtained with wild-type HNF-1α-transfected samples in (A) and those obtained with vector transfected samples in (B). The corresponding 100% values for the numbers with an asterisk are the HNF-1α immunoprecipitated from HNF-1α-transfected cells in (A) and the α-acetyl H3 immunoprecipitated from vector-transfected cells in (B). Errors correspond to deviations obtained from at least two independent experiments. Since the basal levels of acetylated H3 and H4 co-precipitating DNAs were also observed in non-transfected cells (data not shown), taking into account the efficiency of transfection (∼5%), the percentage increases observed in wild-type HNF-1α-transfected samples are clearly underestimates.
Similar articles
- Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins.
Soutoglou E, Papafotiou G, Katrakili N, Talianidis I. Soutoglou E, et al. J Biol Chem. 2000 Apr 28;275(17):12515-20. doi: 10.1074/jbc.275.17.12515. J Biol Chem. 2000. PMID: 10777539 - Transcription factor-specific requirements for coactivators and their acetyltransferase functions.
Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Korzus E, et al. Science. 1998 Jan 30;279(5351):703-7. doi: 10.1126/science.279.5351.703. Science. 1998. PMID: 9445475 - CBP and p300: HATs for different occasions.
Kalkhoven E. Kalkhoven E. Biochem Pharmacol. 2004 Sep 15;68(6):1145-55. doi: 10.1016/j.bcp.2004.03.045. Biochem Pharmacol. 2004. PMID: 15313412 Review. - Versatile molecular glue. Transcriptional control.
Janknecht R, Hunter T. Janknecht R, et al. Curr Biol. 1996 Aug 1;6(8):951-4. doi: 10.1016/s0960-9822(02)00636-x. Curr Biol. 1996. PMID: 8805328 Review.
Cited by
- Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth.
Tronche F, Opherk C, Moriggl R, Kellendonk C, Reimann A, Schwake L, Reichardt HM, Stangl K, Gau D, Hoeflich A, Beug H, Schmid W, Schütz G. Tronche F, et al. Genes Dev. 2004 Mar 1;18(5):492-7. doi: 10.1101/gad.284704. Genes Dev. 2004. PMID: 15037546 Free PMC article. - Co-operation of the transcription factor hepatocyte nuclear factor-4 with Sp1 or Sp3 leads to transcriptional activation of the human haem oxygenase-1 gene promoter in a hepatoma cell line.
Takahashi S, Matsuura N, Kurokawa T, Takahashi Y, Miura T. Takahashi S, et al. Biochem J. 2002 Nov 1;367(Pt 3):641-52. doi: 10.1042/BJ20020819. Biochem J. 2002. PMID: 12133007 Free PMC article. - Enhanced gene activation by Notch and BMP signaling cross-talk.
Takizawa T, Ochiai W, Nakashima K, Taga T. Takizawa T, et al. Nucleic Acids Res. 2003 Oct 1;31(19):5723-31. doi: 10.1093/nar/gkg778. Nucleic Acids Res. 2003. PMID: 14500836 Free PMC article. - Combination Targeting of the Bromodomain and Acetyltransferase Active Site of p300/CBP.
Zucconi BE, Makofske JL, Meyers DJ, Hwang Y, Wu M, Kuroda MI, Cole PA. Zucconi BE, et al. Biochemistry. 2019 Apr 23;58(16):2133-2143. doi: 10.1021/acs.biochem.9b00160. Epub 2019 Apr 11. Biochemistry. 2019. PMID: 30924641 Free PMC article. - Transcription Control of Liver Development.
Tachmatzidi EC, Galanopoulou O, Talianidis I. Tachmatzidi EC, et al. Cells. 2021 Aug 8;10(8):2026. doi: 10.3390/cells10082026. Cells. 2021. PMID: 34440795 Free PMC article. Review.
References
- Ait-Si-Ali S. et al. (1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. - PubMed
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. - PubMed
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. - PubMed
- Frayling T.M. et al. (1997) Mutations in the hepatocyte nuclear factor-1α gene are a common cause of maturity-onset diabetes of the young in the UK. Diabetes, 46, 720–725. - PubMed
- Glucksmann M.A. et al. (1997) Novel mutations and a mutational hotspot in the MODY3 gene. Diabetes, 46, 1081–1086. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical