Arabidopsis genes encoding components of the chloroplastic protein import apparatus - PubMed (original) (raw)
Review
Arabidopsis genes encoding components of the chloroplastic protein import apparatus
D Jackson-Constan et al. Plant Physiol. 2001 Apr.
Abstract
The process of protein import into plastids has been studied extensively using isolated pea (Pisum sativum) chloroplasts. As a consequence, virtually all of the known components of the proteinaceous apparatus that mediates import were originally cloned from pea. With the recent completion of the Arabidopsis genome sequencing project, it is now possible to identify putative homologs of the import components in this species. Our analysis has revealed that Arabidopsis homologs with high sequence similarity exist for all of the pea import complex subunits, making Arabidopsis a valid model for further study of this system. Multiple homologs can be identified for over one-half of the components. In all but one case it is known that more than one of the putative isoforms for a particular subunit are expressed. Thus, it is possible that multiple types of import complexes are present within the same cell, each having a unique affinity for different chloroplastic precursor proteins, depending upon the exact mix of isoforms it contains. Sequence analysis of the putative Arabidopsis homologs for the chloroplast protein import apparatus has revealed many questions concerning subunit function and evolution. It should now be possible to use the genetic tools available in Arabidopsis, including the generation of knockout mutants and antisense technology, to address these questions and learn more about the molecular functions of each of the components during the import process.
Figures
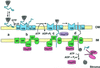
Figure 1
Current model describing the process of protein import into pea chloroplasts. Nuclear-encoded chloroplastic proteins are initially synthesized in the cytoplasm with a transit peptide (teal) that targets them to the plastid surface (a). In a process stimulated by GTP, the precursor protein associates with the components (blue) of the outer envelope membrane translocon (b). Hydrolysis of ATP in the cytoplasm and/or intermembrane space causes the precursor to interact with the components (green) of the inner membrane translocon as well (c). It is postulated that this step may be assisted by chaperones residing in the intermembrane space (purple). Hydrolysis of stromal ATP results in the complete translocation of the precursor protein into the chloroplast interior, where the transit peptide is removed (d). This final step is mediated at least in part by stromal factors (red). The numbers within the components of the outer and inner membrane translocons refer to the calculated molecular mass of each subunit. OM, Outer membrane; IM, inner membrane.
Figure 2
Multiple sequence alignment for the putative chloroplast-localized Arabidopsis Hsp70 isoforms. Shaded residues designate sequence identities between two or more of the proteins. The predicted transit peptide is indicated (>). The predicted cleavage site is based on sequence identity to a pea chloroplastic Hsp70 (accession no. L03299) and has not been experimentally verified. The alignment was created using the PileUp program from the Wisconsin package of sequence analysis tools (Genetics Computer Group, Madison, WI).
Similar articles
- Pea chloroplast DnaJ-J8 and Toc12 are encoded by the same gene and localized in the stroma.
Chiu CC, Chen LJ, Li HM. Chiu CC, et al. Plant Physiol. 2010 Nov;154(3):1172-82. doi: 10.1104/pp.110.161224. Epub 2010 Sep 14. Plant Physiol. 2010. PMID: 20841453 Free PMC article. - Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts.
Su PH, Li HM. Su PH, et al. Plant Cell. 2010 May;22(5):1516-31. doi: 10.1105/tpc.109.071415. Epub 2010 May 18. Plant Cell. 2010. PMID: 20484004 Free PMC article. - A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway.
Tranel PJ, Froehlich J, Goyal A, Keegstra K. Tranel PJ, et al. EMBO J. 1995 Jun 1;14(11):2436-46. doi: 10.1002/j.1460-2075.1995.tb07241.x. EMBO J. 1995. PMID: 7781598 Free PMC article. - How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts.
Macasev D, Newbigin E, Whelan J, Lithgow T. Macasev D, et al. Plant Physiol. 2000 Jul;123(3):811-6. doi: 10.1104/pp.123.3.811. Plant Physiol. 2000. PMID: 10889230 Free PMC article. Review. No abstract available. - Structural considerations of folded protein import through the chloroplast TOC/TIC translocons.
Ganesan I, Theg SM. Ganesan I, et al. FEBS Lett. 2019 Mar;593(6):565-572. doi: 10.1002/1873-3468.13342. Epub 2019 Mar 5. FEBS Lett. 2019. PMID: 30775779 Review.
Cited by
- Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences.
Durnford DG, Gray MW. Durnford DG, et al. Eukaryot Cell. 2006 Dec;5(12):2079-91. doi: 10.1128/EC.00222-06. Epub 2006 Sep 22. Eukaryot Cell. 2006. PMID: 16998072 Free PMC article. - Common ground for protein translocation: access control for mitochondria and chloroplasts.
Schleiff E, Becker T. Schleiff E, et al. Nat Rev Mol Cell Biol. 2011 Jan;12(1):48-59. doi: 10.1038/nrm3027. Epub 2010 Dec 8. Nat Rev Mol Cell Biol. 2011. PMID: 21139638 Review. - Import of Soluble Proteins into Chloroplasts and Potential Regulatory Mechanisms.
Sjuts I, Soll J, Bölter B. Sjuts I, et al. Front Plant Sci. 2017 Feb 8;8:168. doi: 10.3389/fpls.2017.00168. eCollection 2017. Front Plant Sci. 2017. PMID: 28228773 Free PMC article. Review. - New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways.
Fish M, Nash D, German A, Overton A, Jelokhani-Niaraki M, Chuong SDX, Smith MD. Fish M, et al. Int J Mol Sci. 2022 Jan 29;23(3):1571. doi: 10.3390/ijms23031571. Int J Mol Sci. 2022. PMID: 35163495 Free PMC article. Review. - Modifications at the A-domain of the chloroplast import receptor Toc159.
Agne B, Kessler F. Agne B, et al. Plant Signal Behav. 2010 Nov;5(11):1513-6. doi: 10.4161/psb.5.11.13707. Epub 2010 Nov 1. Plant Signal Behav. 2010. PMID: 21057194 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials