An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast - PubMed (original) (raw)
An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast
R M Long et al. J Cell Biol. 2001.
Abstract
The localization of ASH1 mRNA to the distal tip of budding yeast cells is essential for the proper regulation of mating type switching in Saccharomyces cerevisiae. A localization element that is predominantly in the 3'-untranslated region (UTR) can direct this mRNA to the bud. Using this element in the three-hybrid in vivo RNA-binding assay, we identified a protein, Loc1p, that binds in vitro directly to the wild-type ASH1 3'-UTR RNA, but not to a mutant RNA incapable of localizing to the bud nor to several other mRNAs. LOC1 codes for a novel protein that recognizes double-stranded RNA structures and is required for efficient localization of ASH1 mRNA. Accordingly, Ash1p gets symmetrically distributed between daughter and mother cells in a loc1 strain. Surprisingly, Loc1p was found to be strictly nuclear, unlike other known RNA-binding proteins involved in mRNA localization which shuttle between the nucleus and the cytoplasm. We propose that efficient cytoplasmic ASH1 mRNA localization requires a previous interaction with specific nuclear factors.
Figures
Figure 1
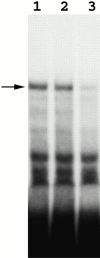
A band mobility shift assay identifies proteins that specifically bind the E3 element. Radiolabeled E3 transcripts were incubated with yeast protein extracts, and then with RNase T1 and heparin at room temperature. RNA–protein complexes were resolved in 4% native gel and visualized by autoradiography. Lane 1, E3 transcripts incubated with K699 extract; lanes 2 and 3, E3 transcripts incubated with K699 extracts in the presence of 200× molar excess of pGEM RNA and unlabeled E3 transcripts, respectively.
Figure 3
The formation of the specific RNA–protein complex is dependent on Loc1p. Band mobility shift assays were performed as mentioned in the legend to Fig. 1. Lane 1, E3 RNA probe only, no protein extract. Lanes 2 and 3, E3 transcripts were incubated with yeast extracts of YLM090 (loc1) and K699 (wild-type) strains. Lane 4, E3 transcripts incubated with extracts of YLM090 (loc1) complemented with a LOC1-myc plasmid.
Figure 2
Amino acid sequence of Loc1p as predicted from the Yeast Genome Sequencing project. One remarkable feature of Loc1p is the abundance of arginine and lysine in this protein. Loc1p contains 20.1% lysine and 9.8% arginine. The pI value of 10.79 for Loc1p is a reflection of the abundance of positive charge in this protein.
Figure 5
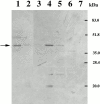
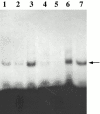
Affinity purification of Loc1p-myc from an E3 RNA column. RNA affinity-purified fractions were analyzed by Western blot with anti-myc antibody. Lane 1, starting extracts; lane 2, flowthrough fraction; lane 3, wash fraction; lane 4, elution fraction with high salt; lanes 5 and 6, elution fractions from RNA affinity columns using M9 and pGEM 3Z RNA as ligands, respectively; and lane 7, protein extract from strain K699 (no myc-tagged protein). Arrow, Loc1p-myc protein.
Figure 4
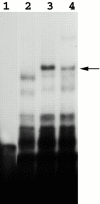
Recombinant Loc1p-GST fusion binds to E3. Lanes 1 and 2, E3 transcripts incubated with bacterial extracts containing recombinant GST-Loc1p but immunodepleted by GST antibody or glutathione beads, respectively. Lane 3, E3 transcripts incubated with the extract before immunodepletion. Lanes 4, 5, and 6, E3 transcripts incubated with bacterial extract containing recombinant GST-Loc1p in the presence of 100× excess of unlabeled E3, IRE, and pGEM 3Z RNAs, respectively. Lane 7, E3 transcripts incubated with SDS-PAGE purified recombinant Loc1p-GST fusion. The arrow indicates the E3/Loc1p–GST complex.
Figure 7
The ASH1 mRNA is delocalized in a loc1 yeast strain. (A) Fluorescent in situ hybridization for ASH1 mRNA in wild-type (K699) and loc1 (YLM090) yeast cells. ASH1 mRNA is expressed from a multicopy plasmid (YEPlac181). DAPI, DNA staining; NOM., Nomarski. The ASH1 mRNA is tightly localized at the bud tip of a wild-type cell, whereas it is delocalized in the cytoplasm of a late anaphase loc1 yeast cell. Bar, 10 μm. (B) Immunofluorescence detection of the Ash1p-myc in postanaphase YML094 (cla4) and YML093 (cla4 loc1) yeast cells. DAPI, DNA staining; NOM., Nomarski. Note the elongated bud neck caused by the deletion of the CLA4 gene. In the majority of cells, Ash1p-myc is symmetrically distributed in the loc1 strain. Bar, 10 μm.
Figure 6
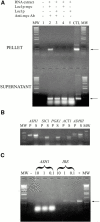
ASH1 mRNA coimmunoprecipitates specifically with Loc1p-myc. (A) RT-PCR amplification of ASH1 mRNA extracted from pellet (top) or supernatant (bottom) after immunoprecipitation. Lane 1, RT-PCR reaction without RNA extract. Lanes 2 and 4, immunoprecipitation from strain expressing Loc1p-myc with (lane 2) or without (lane 4) anti-myc antibody. Lanes 3 and 5, immunoprecipitation from strain expressing Loc1p with (lane 3) or without (lane 5) anti-myc antibody. CTL, control PCR amplification of the ASH1 gene. MW, molecular weight marker. Arrows indicate the ASH1 PCR fragment of 375 bp. (B) RT-PCR amplifications of various mRNAs after immunoprecipitation of Loc1p-myc. P, RNA extracted from the pellet; S, RNA extracted from the supernatant; MW, molecular weight marker. (C) Competition of the immunoprecipitation of ASH1 mRNA by the localization elements. Yeast extract was incubated with either 10, 1, and 0.1 μg of an equimolar mix of the four ASH1 localization elements (ASH1) or the IRE RNA (IRE) before immunoprecipitation of the Loc1p-myc protein and RT-PCR of the ASH1 mRNA. MW, molecular weight marker; −, negative control (no yeast extract); +, positive control (IP of ASH1 mRNA and RT-PCR without competition).
Figure 8
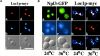
Intracellular distribution of the Loc1p-myc protein. (A) Immunofluorescence detection of the Loc1p-myc. Strain YLM090 (loc1) was transformed with the plasmid pRL094 (LOC1-MYC). Loc1p-myc (in red) is expressed throughout the cell cycle and appears to be strictly nuclear (see DAPI, in blue). The two panels show cells before (left) and during (right) anaphase. Bar, 10 μm. (B) Shuttling assay for Loc1p-myc protein. (a–f) Npl3p-GFP at 24°C (a–c) and 36°C (d–f). (g–l) Loc1p-myc at 24°C (g–i) and 36°C (j–l). Whereas the Npl3p-GFP (in green) accumulates in the cytoplasm after a temperature shift from 24°C (a) to 36°C (d), Loc1p-myc (in red) remains in the nucleus at 24°C (g) and 36°C (j). g and j are an overlap of images h and i, and k and l, respectively. b, e, i, and l are DAPI staining (in blue) and c and f are Nomarski. Bar, 10 μm. (C) Heterokaryon shuttling assay for Loc1p-myc. (a–c) Loc1p-myc. (d–f) Npl3p-GFP. Binuclear yeasts show no accumulation of Loc1p-myc (in red) in the second nucleus (a), whereas Npl3p-GFP (in green) appears in both nuclei after cell fusion (d). b and e are DAPI staining (in blue) and c and f are Nomarski. Bar, 10 μm.
Figure 8
Intracellular distribution of the Loc1p-myc protein. (A) Immunofluorescence detection of the Loc1p-myc. Strain YLM090 (loc1) was transformed with the plasmid pRL094 (LOC1-MYC). Loc1p-myc (in red) is expressed throughout the cell cycle and appears to be strictly nuclear (see DAPI, in blue). The two panels show cells before (left) and during (right) anaphase. Bar, 10 μm. (B) Shuttling assay for Loc1p-myc protein. (a–f) Npl3p-GFP at 24°C (a–c) and 36°C (d–f). (g–l) Loc1p-myc at 24°C (g–i) and 36°C (j–l). Whereas the Npl3p-GFP (in green) accumulates in the cytoplasm after a temperature shift from 24°C (a) to 36°C (d), Loc1p-myc (in red) remains in the nucleus at 24°C (g) and 36°C (j). g and j are an overlap of images h and i, and k and l, respectively. b, e, i, and l are DAPI staining (in blue) and c and f are Nomarski. Bar, 10 μm. (C) Heterokaryon shuttling assay for Loc1p-myc. (a–c) Loc1p-myc. (d–f) Npl3p-GFP. Binuclear yeasts show no accumulation of Loc1p-myc (in red) in the second nucleus (a), whereas Npl3p-GFP (in green) appears in both nuclei after cell fusion (d). b and e are DAPI staining (in blue) and c and f are Nomarski. Bar, 10 μm.
Similar articles
- Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p.
Shen Z, Paquin N, Forget A, Chartrand P. Shen Z, et al. Mol Biol Cell. 2009 Apr;20(8):2265-75. doi: 10.1091/mbc.e08-11-1151. Epub 2009 Feb 25. Mol Biol Cell. 2009. PMID: 19244342 Free PMC article. - She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p.
Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. Böhl F, et al. EMBO J. 2000 Oct 16;19(20):5514-24. doi: 10.1093/emboj/19.20.5514. EMBO J. 2000. PMID: 11032818 Free PMC article. - Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo.
Chartrand P, Meng XH, Singer RH, Long RM. Chartrand P, et al. Curr Biol. 1999 Mar 25;9(6):333-6. doi: 10.1016/s0960-9822(99)80144-4. Curr Biol. 1999. PMID: 10209102 - RNA localization in yeast: moving towards a mechanism.
Gonsalvez GB, Urbinati CR, Long RM. Gonsalvez GB, et al. Biol Cell. 2005 Jan;97(1):75-86. doi: 10.1042/BC20040066. Biol Cell. 2005. PMID: 15601259 Review. - Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast.
Niedner A, Edelmann FT, Niessing D. Niedner A, et al. RNA Biol. 2014;11(8):998-1009. doi: 10.4161/rna.29946. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482892 Free PMC article. Review.
Cited by
- Linking transport and translation of mRNAs with endosomes and mitochondria.
Müntjes K, Devan SK, Reichert AS, Feldbrügge M. Müntjes K, et al. EMBO Rep. 2021 Oct 5;22(10):e52445. doi: 10.15252/embr.202152445. Epub 2021 Aug 17. EMBO Rep. 2021. PMID: 34402186 Free PMC article. Review. - Intracellular mRNA transport and localized translation.
Das S, Vera M, Gandin V, Singer RH, Tutucci E. Das S, et al. Nat Rev Mol Cell Biol. 2021 Jul;22(7):483-504. doi: 10.1038/s41580-021-00356-8. Epub 2021 Apr 9. Nat Rev Mol Cell Biol. 2021. PMID: 33837370 Free PMC article. Review. - Puf6 and Loc1 Are the Dedicated Chaperones of Ribosomal Protein Rpl43 in Saccharomyces cerevisiae.
Liang KJ, Yueh LY, Hsu NH, Lai JS, Lo KY. Liang KJ, et al. Int J Mol Sci. 2019 Nov 26;20(23):5941. doi: 10.3390/ijms20235941. Int J Mol Sci. 2019. PMID: 31779129 Free PMC article. - Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.
Ahn HW, Tucker JM, Arribere JA, Garfinkel DJ. Ahn HW, et al. Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article. - The Roles of Puf6 and Loc1 in 60S Biogenesis Are Interdependent, and Both Are Required for Efficient Accommodation of Rpl43.
Yang YT, Ting YH, Liang KJ, Lo KY. Yang YT, et al. J Biol Chem. 2016 Sep 9;291(37):19312-23. doi: 10.1074/jbc.M116.732800. Epub 2016 Jul 25. J Biol Chem. 2016. PMID: 27458021 Free PMC article.
References
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, ; Cold Spring Harbor, New York: 1997.
- Beach D.L., Salmon E.D., Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. - PubMed
- Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. - PubMed
- Bobola N., Jansen R.-P., Shin T.H., Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM057071/GM/NIGMS NIH HHS/United States
- R01 GM060392/GM/NIGMS NIH HHS/United States
- F32 HD008088/HD/NICHD NIH HHS/United States
- F32 HD08088/HD/NICHD NIH HHS/United States
- 2-T32-CA09475/CA/NCI NIH HHS/United States
- T32 CA009475/CA/NCI NIH HHS/United States
- GM57071/GM/NIGMS NIH HHS/United States
- GM60392/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases