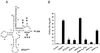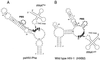Identification of critical elements in the tRNA acceptor stem and T(Psi)C loop necessary for human immunodeficiency virus type 1 infectivity - PubMed (original) (raw)
Identification of critical elements in the tRNA acceptor stem and T(Psi)C loop necessary for human immunodeficiency virus type 1 infectivity
Q Yu et al. J Virol. 2001 May.
Abstract
A mutant human immunodeficiency virus type 1 (HIV-1) with a primer binding site (PBS) complementary to yeast tRNA(Phe) (psHIV-Phe), which relies on exogenous yeast tRNA(Phe) as reverse transcription primer, was used to investigate elements in the tRNA acceptor stem and T(Psi)C stem-loop required for the tRNA primer selection and use in HIV-1 replication. tRNA(Phe) mutants with two- or four-nucleotide deletions in the 3' end retained the capacity to complement replication of psHIV-Phe. tRNA(Phe) mutants with an extended 5' end had reduced capacity for complementation, which could be restored by extension of the 3' end of these tRNA(Phe) mutants with sequences complementary to the HIV-1 U5 region. Further analysis of mutations in the acceptor stem of tRNA(Phe) suggested that an intact acceptor stem RNA structure is important for complementation. Analysis of single-nucleotide changes in the T(Psi)C stem-loop of tRNA(Phe) revealed an unexpected, essential role of this region for rescue of psHIV-Phe.
Figures
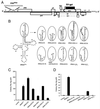
FIG. 1
Effect of tRNAPhe acceptor stem mutations on complementation of psHIV-Phe. (A) psHIV-Phe proviral genome. The defective psHIV-Phe proviral genome contains a PBS complementary to yeast tRNAPhe (PBSPhe). The env gene of HIV was substituted by the gpt gene under the control of simian virus 40 early promoter (SV-gpt). To generate pseudoviruses, this plasmid was cotransfected with the plasmid encoding vesicular stomatitis virus G protein either with or without tRNA. Successful infection of cells with the pseudoviruses confers resistance to mycophenolic acid. (B) Illustration of tRNAPhe mutants with nucleotide changes at the acceptor stem. The added or substituted nucleotides are depicted in bold. Only the terminal portion of each tRNAPhe mutant (circled) is illustrated. (C and D) Numbers of drug-resistant colonies derived from infection of the psHIV-Phe pseudoviruses complemented by indicated mutants. Values are means of data obtained from three independent experiments, with standard deviations depicted (error bars).
FIG. 2
Mutations in the tRNAPhe TΨC stem-loop affect complementation of psHIV-Phe. (A) Illustrations of the positions of the nucleotides mutated in designated tRNAPhe mutants. (B) The psHIV-Phe rescue capacity of the tRNAPhe mutants as measured by drug-resistant colony numbers. Values are means of data obtained from three independent experiments, with standard deviations depicted (error bars).
FIG. 3
RNA secondary structures of the PBS and surrounding regions in psHIV-Phe (A) and in wild-type HIV-1 (HXB2 strain) (B) predicted by the M-fold structure modeling program. The PBS regions are in bold. The boxed nucleotides in a loop region represent the PBS-downstream sequence complementary to the TψC loop sequence of tRNAs. Potential loop-loop interactions between the tRNA TΨC loop and the PBS-downstream sequence of the viral RNA genome are depicted. The drawings are for illustration only and are not to scale.
Similar articles
- Selection of retroviral reverse transcription primer is coordinated with tRNA biogenesis.
Kelly NJ, Palmer MT, Morrow CD. Kelly NJ, et al. J Virol. 2003 Aug;77(16):8695-701. doi: 10.1128/jvi.77.16.8695-8701.2003. J Virol. 2003. PMID: 12885888 Free PMC article. - Essential regions of the tRNA primer required for HIV-1 infectivity.
Yu Q, Morrow CD. Yu Q, et al. Nucleic Acids Res. 2000 Dec 1;28(23):4783-9. doi: 10.1093/nar/28.23.4783. Nucleic Acids Res. 2000. PMID: 11095691 Free PMC article. - Yeast tRNA(Phe) expressed in human cells can be selected by HIV-1 for use as a reverse transcription primer.
Kelly NJ, Morrow CD. Kelly NJ, et al. Virology. 2003 Sep 1;313(2):354-63. doi: 10.1016/s0042-6822(03)00243-5. Virology. 2003. PMID: 12954204 - Ribosomal binding of modified tRNA anticodons related to thermal stability.
Ashraf SS, Guenther R, Ye W, Lee Y, Malkiewicz A, Agris PF. Ashraf SS, et al. Nucleic Acids Symp Ser. 1997;(36):58-60. Nucleic Acids Symp Ser. 1997. PMID: 9478206 Review.
Cited by
- Structural elements of the tRNA TPsiC loop critical for nucleocytoplasmic transport are important for human immunodeficiency virus type 1 primer selection.
Kelly NJ, Morrow CD. Kelly NJ, et al. J Virol. 2005 May;79(10):6532-9. doi: 10.1128/JVI.79.10.6532-6539.2005. J Virol. 2005. PMID: 15858038 Free PMC article. - Selection of retroviral reverse transcription primer is coordinated with tRNA biogenesis.
Kelly NJ, Palmer MT, Morrow CD. Kelly NJ, et al. J Virol. 2003 Aug;77(16):8695-701. doi: 10.1128/jvi.77.16.8695-8701.2003. J Virol. 2003. PMID: 12885888 Free PMC article. - Mutations in the TPsiC loop of E. coli tRNALys,3 have varied effects on in trans complementation of HIV-1 replication.
Yu W, McCulley A, Morrow CD. Yu W, et al. Virol J. 2007 Jan 11;4:5. doi: 10.1186/1743-422X-4-5. Virol J. 2007. PMID: 17217532 Free PMC article. - Complementation of human immunodeficiency virus type 1 replication by intracellular selection of Escherichia coli formula supplied in trans.
McCulley A, Morrow CD. McCulley A, et al. J Virol. 2006 Oct;80(19):9641-50. doi: 10.1128/JVI.00709-06. J Virol. 2006. PMID: 16973568 Free PMC article. - Nucleotides within the anticodon stem are important for optimal use of tRNA(Lys,3) as the primer for HIV-1 reverse transcription.
McCulley A, Morrow CD. McCulley A, et al. Virology. 2007 Jul 20;364(1):169-77. doi: 10.1016/j.virol.2007.02.010. Epub 2007 Mar 21. Virology. 2007. PMID: 17368706 Free PMC article.
References
- Gregoire C J, Gautheret D, Loret E P. No tRNA
unwinding in a complex with HIV NCp7. J Biol Chem. 1997;272:25143–25148. - PubMed
- Gregoire C J, Gautheret D, Loret E P. No tRNA
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI027767/AI/NIAID NIH HHS/United States
- GM56544/GM/NIGMS NIH HHS/United States
- AI34749/AI/NIAID NIH HHS/United States
- AI-27767/AI/NIAID NIH HHS/United States
- R37 AI034749/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases