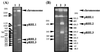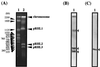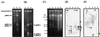Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1 - PubMed (original) (raw)
Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1
S Shimizu et al. Appl Environ Microbiol. 2001 May.
Abstract
A strong polychlorinated biphenyl (PCB) degrader, Rhodococcus sp. strain RHA1, has diverse biphenyl/PCB degradative genes and harbors huge linear plasmids, including pRHL1 (1,100 kb), pRHL2 (450 kb), and pRHL3 (330 kb). The diverse degradative genes are distributed mainly on the pRHL1 and pRHL2 plasmids. In this study, the structural and functional characteristics of pRHL2 were determined. We constructed a physical map of pRHL2, and the degradative enzyme genes, including bphB2, etbD2, etbC, bphDEF, bphC2, and bphC4, were localized in three regions. Conjugal transfer of pRHL2 between RHA1 mutant derivatives was observed at a frequency of 7.5 x 10(-5) transconjugant per recipient. These results suggested that the linear plasmid is a possible determinant of propagation of the diverse degradative genes in rhodococci. The termini of pRHL2 were cloned and sequenced. The left and right termini of pRHL2 had 3-bp perfect terminal inverted repeats and were not as similar to each other (64% identity) as the known actinomycete linear replicons are. Southern hybridization analysis with pRHL2 terminal probes suggested that the right terminus of pRHL2 is similar to pRHL1 and pRHL3 termini. Retardation of both terminal fragments in the gel shift assay indicated that each terminus of pRHL2 is linked to a protein. We suggest that pRHL2 has invertron termini, as has been reported previously for Streptomyces linear replicons.
Figures
FIG. 1
PFGE of RHA1 linear plasmids performed with long (A) and short (B) pulse times. The positions of the chromosome and linear plasmids are indicated. (A) The pulse time was increased from 60 to 90 s during 23 h of electrophoresis. The voltage was adjusted to 6 V/cm. (B) The pulse time was increased from 20 to 30 s during 23 h of electrophoresis. The voltage was adjusted to 6 V/cm. Lane 1, S. cerevisiae chromosome marker, lane 2, total DNA of RHA1; lane 3, λ ladder marker.
FIG. 2
Physical map of pRHL2 and localization of degradative genes. The locations of degradative genes are indicated by bars below the map. The insert region of each pRHL2 subclone is also indicated below the map.
FIG. 3
Linear plasmids in transconjugants. Linear plasmids of a donor (RCA1), a recipient (RDO5), and transconjugants were separated by PFGE. The pulse time was increased from 60 to 90 s during 23 h of electrophoresis. The voltage was adjusted to 6 V/cm. Lane 1, S. cerevisiae chromosome marker; lane 2, RCA1; lane 3, RDO5; lanes 4 to 10, RDO5 transconjugants.
FIG. 4
Alignment of the terminal sequences of pRHL2 and pHG201. The dashes indicate gaps; the asterisks indicate identical nucleotide residues in pRHL2 and pHG201. The 3-bp perfect terminal inverted repeats are indicated by boldface type. The inverted repeats of pHG201 containing the control motif GCTXCGC which were identified by Kalkus et al. (11) are indicated by converging arrows.
FIG. 5
Southern hybridization of linear plasmids of RHA1 with the terminus probes of pRHL2. (A) Results of PFGE. The sizes of marker fragments and the positions of the chromosome and linear plasmids are indicated on the left and right, respectively. The pulse time was increased from 60 to 90 s during 23 h of electrophoresis. The voltage was adjusted to 6 V/cm. Lane 1, S. cerevisiae chromosome marker; lane 2, linear plasmids of RHA1. (B and C) Results of hybridization performed with the right (B) and left (C) end probes. The 1.1-kb _Eco_RI and 0.45-kb _Eco_RI-_Sal_I fragments of pRHL2 were used as the right and left end probes, respectively. The lanes contained linear plasmids of RHA1. The bands of linear plasmids are indicated by arrowheads.
FIG. 6
(A and B) Normal PFGE (A) and SDS-PFGE (B) of linear plasmid DNAs obtained with and without proteinase K treatment. The pulse time was increased from 60 to 90 s during 23 h of electrophoresis. The voltage was adjusted to 6 V/cm. Lane 1, S. cerevisiae chromosome marker; lane 2, proteinase K-treated plasmid DNA; lane 3, non-proteinase K-treated plasmid DNA. The positions of the chromosome and plasmids are indicated on the left and right. (C to E) Results of PFGE (C) and hybridization (D and E) of total restriction fragments obtained with and without proteinase K treatment. Hybridization was performed with the right (D) and left (E) end probes. The 1.1-kb _Eco_RI and 0.45-kb _Eco_RI-_Sal_I fragments of pRHL2 were used as the right and left end probes, respectively. The pulse time was increased from 3 to 12 s during 21 h of electrophoresis. The voltage was adjusted to 5.1 V/cm. The sizes of marker fragments in panel C are indicated on the left. The bands of interest in panels D and E are indicated by solid arrowheads. Minor bands in panel D which seemed to be derived from the termini of pRHL1 and pRHL3 are indicated by open arrowheads. Lane 1, non-proteinase K-treated _Ase_I digests; lane 2, proteinase K-treated _Ase_I digests; lane 3, λ ladder plus _Hin_dIII-digested λ DNA markers; lane 4, non-proteinase K-treated _Hpa_I digests; lane 5, proteinase K-treated _Hpa_I digests.
Similar articles
- Functional characterization of a catabolic plasmid from polychlorinated- biphenyl-degrading Rhodococcus sp. strain RHA1.
Warren R, Hsiao WW, Kudo H, Myhre M, Dosanjh M, Petrescu A, Kobayashi H, Shimizu S, Miyauchi K, Masai E, Yang G, Stott JM, Schein JE, Shin H, Khattra J, Smailus D, Butterfield YS, Siddiqui A, Holt R, Marra MA, Jones SJ, Mohn WW, Brinkman FS, Fukuda M, Davies J, Eltis LD. Warren R, et al. J Bacteriol. 2004 Nov;186(22):7783-95. doi: 10.1128/JB.186.22.7783-7795.2004. J Bacteriol. 2004. PMID: 15516593 Free PMC article. - The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1.
Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild JE, Hatta T, Kimbara K, Yano K, Fukuda M. Masai E, et al. Gene. 1997 Mar 10;187(1):141-9. doi: 10.1016/s0378-1119(96)00748-2. Gene. 1997. PMID: 9073078 - Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1.
Yamada A, Kishi H, Sugiyama K, Hatta T, Nakamura K, Masai E, Fukuda M. Yamada A, et al. Appl Environ Microbiol. 1998 Jun;64(6):2006-12. doi: 10.1128/AEM.64.6.2006-2012.1998. Appl Environ Microbiol. 1998. PMID: 9603807 Free PMC article. - [PCB degradation systems in Rhodococcus bacteria: multiplex enzyme system determined by multiple isozyme genes].
Fukuda M, Miyauchi K, Masai E. Fukuda M, et al. Tanpakushitsu Kakusan Koso. 2005 Oct;50(12):1541-7. Tanpakushitsu Kakusan Koso. 2005. PMID: 16218454 Review. Japanese. No abstract available. - Biotechnological Potential of Rhodococcus Biodegradative Pathways.
Kim D, Choi KY, Yoo M, Zylstra GJ, Kim E. Kim D, et al. J Microbiol Biotechnol. 2018 Jul 28;28(7):1037-1051. doi: 10.4014/jmb.1712.12017. J Microbiol Biotechnol. 2018. PMID: 29913546 Review.
Cited by
- New insights into the genome of Rhodococcus ruber strain Chol-4.
Guevara G, Castillo Lopez M, Alonso S, Perera J, Navarro-Llorens JM. Guevara G, et al. BMC Genomics. 2019 May 2;20(1):332. doi: 10.1186/s12864-019-5677-2. BMC Genomics. 2019. PMID: 31046661 Free PMC article. - Strategies and approaches in plasmidome studies-uncovering plasmid diversity disregarding of linear elements?
Dib JR, Wagenknecht M, Farías ME, Meinhardt F. Dib JR, et al. Front Microbiol. 2015 May 26;6:463. doi: 10.3389/fmicb.2015.00463. eCollection 2015. Front Microbiol. 2015. PMID: 26074886 Free PMC article. Review. - Novel Multidrug-Resistant Enterococcal Mobile Linear Plasmid pELF1 Encoding vanA and vanM Gene Clusters From a Japanese Vancomycin-Resistant Enterococci Isolate.
Hashimoto Y, Taniguchi M, Uesaka K, Nomura T, Hirakawa H, Tanimoto K, Tamai K, Ruan G, Zheng B, Tomita H. Hashimoto Y, et al. Front Microbiol. 2019 Nov 13;10:2568. doi: 10.3389/fmicb.2019.02568. eCollection 2019. Front Microbiol. 2019. PMID: 31798546 Free PMC article. - Different Roles of Dioxin-Catabolic Plasmids in Growth, Biofilm Formation, and Metabolism of Rhodococcus sp. Strain p52.
Wang X, Wu Y, Chen M, Fu C, Xu H, Li L. Wang X, et al. Microorganisms. 2024 Aug 17;12(8):1700. doi: 10.3390/microorganisms12081700. Microorganisms. 2024. PMID: 39203542 Free PMC article. - Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites.
Vaillancourt FH, Haro MA, Drouin NM, Karim Z, Maaroufi H, Eltis LD. Vaillancourt FH, et al. J Bacteriol. 2003 Feb;185(4):1253-60. doi: 10.1128/JB.185.4.1253-1260.2003. J Bacteriol. 2003. PMID: 12562795 Free PMC article.
References
- Fukuda M, Shimizu S, Okita N, Seto M, Masai E. Structural alteration of linear plasmids encoding the genes for polychlorinated biphenyl degradation in Rhodococcus strain RHA1. Antonie Leeuwenhoek. 1998;74:169–173. - PubMed
- Hashimoto Y, Nishiyama M, Yu F, Watanabe I, Horinouchi S, Beppu T. Development of a host-vector system in a Rhodococcus strain and its use for expression of the cloned nitrile hydratase gene cluster. J Gen Microbiol. 1992;138:1003–1010. - PubMed
- Hatta T, Shimada T, Yoshihara T, Yamada A, Masai E, Fukuda M, Kiyohara H. meta-Fission product hydrolases from a strong PCB degrader Rhodococcus sp. strain RHA1. J Ferment Bioeng. 1998;85:174–179.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources