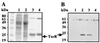Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor - PubMed (original) (raw)
Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor
N Mani et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Clostridium difficile, a causative agent of antibiotic-associated diarrhea and its potentially lethal form, pseudomembranous colitis, produces two large protein toxins that are responsible for the cellular damage associated with the disease. The level of toxin production appears to be critical for determining the severity of the disease, but the mechanism by which toxin synthesis is regulated is unknown. The product of a gene, txeR, that lies just upstream of the tox gene cluster was shown to be needed for tox gene expression in vivo and to activate promoter-specific transcription of the tox genes in vitro in conjunction with RNA polymerases from C. difficile, Bacillus subtilis, or Escherichia coli. TxeR was shown to function as an alternative sigma factor for RNA polymerase. Because homologs of TxeR regulate synthesis of toxins and a bacteriocin in other Clostridium species, TxeR appears to be a prototype for a novel mode of regulation of toxin genes.
Figures
Figure 1
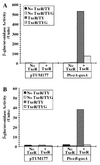
In vivo transcription activation of P_tox_-gusA fusions by txeR in trans in C. perfringens. Fragments of DNA carrying either toxA (A) or_toxB_ (B) gene promoters were cloned in the reporter fusion vector pTUM177 and introduced into C. perfringens with or without the txeR gene in_trans_. β-Glucuronidase activity of late stationary phase cells grown in TY or TYG medium was assayed as described previously (7). In control experiments, cells carrying the fusion vector alone were assayed. Solid bars, TY medium, no TxeR; open bars, TYG medium, no TxeR; striped bars, TY medium with TxeR; dotted bars, TYG medium, with TxeR.
Figure 2
Overproduction and purification of TxeR. (A) SDS/PAGE analysis of protein extracts from E. coli BL21λDE3 (pLysS) carrying either pCD54 or pET28-b. Lanes: 1, crude cell extracts from E. coli carrying the vector pET28-b; 2, crude cell extracts from E. coli carrying pCD54 (expressing TxeR); 3, TxeR-containing fraction from_E. coli_ carrying pCD54 retained and eluted from a Ni+-NTA column; 4, an equivalent protein fraction from crude extract of E. coli carrying the vector pET28-b after Ni+-NTA chromatography. (B) Immunodetection of TxeR by using anti-TxeR antibodies. The protein samples in each lane correspond to those in A. The bands in lanes 2 and 3 corresponding to TxeR are indicated by an arrow.
Figure 3
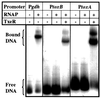
Gel mobility retardation of tox promoters with C. difficile RNA polymerase and TxeR; gel mobility retardation of the C. difficile glutamate dehydrogenase gene promoter (P_gdh_), toxin B gene promoter (P_toxB_), and toxin A gene promoter (P_toxA_) by C. difficile RNA polymerase. For gdh promoter, left lane is P_gdh_ DNA alone and right lane is P_gdh_ DNA incubated with C. difficile RNA polymerase. For toxB and toxA promoters, left lane is tox promoter DNA alone, middle lane is_tox_ promoter DNA with C. difficile RNA polymerase, and right lane is tox promoter DNA with_C. difficile_ RNA polymerase and TxeR, respectively.
Figure 4
In vitro transcription activation from_tox_ promoters by RNA polymerase and TxeR. In vitro run-off transcription reactions were performed by using RNA polymerases from E. coli (Ec), B. subtilis (Bs), or C. difficile (Cd) and DNA fragments containing either the toxA promoter (P_toxA_) or the toxB promoter (P_toxB_) incubated in the absence or presence of partially purified TxeR. A control transcription reaction performed on the gdh promoter DNA with C. difficile RNA polymerase is shown in the leftmost lane. The expected sizes of run-off transcripts (indicated by arrows) are: P_gdh_, 162 nt; P_toxA_, 255 nt; P_toxB_, 175 nt. An additional read-through transcript of 291 nt is made from the_gdh_ promoter.
Figure 5
Gel mobility retardation of tox promoters with E. coli RNA polymerase core enzyme and TxeR. Gel mobility retardation of the C. difficile gdh gene promoter (P_gdh_) (A), the toxA gene promoter (P_toxA_) (B), and_toxB_ gene promoter (P_toxB_) (C) by E. coli RNA polymerase core enzyme alone or in the presence of either TxeR or contaminating proteins (Prot-TxeR) from E. coli not expressing TxeR. The triangles in B and C indicate the use of increasing amounts of TxeR (50, 100, and 200 nM).
Figure 6
In vitro transcription activation from_tox_ promoters by RNA polymerase core enzyme and TxeR.In vitro transcription reactions were carried out in the presence of E. coli RNA polymerase holoenzyme (H) or core enzyme (C) (A and B) or B. subtilis RNA polymerase holoenzyme (H) or core enzyme (C) (C) with or without TxeR by using as templates DNA fragments containing the promoters of the gdh,toxA, or toxB genes. In lanes marked with an asterisk, contaminating proteins from E. coli not expressing TxeR were added. The arrows indicate the positions of the_toxA_ and toxB transcripts. The sizes of expected gdh transcripts are 122 and 251 nt.
Figure 7
Interaction of purified TxeR with E. coli RNA polymerase core enzyme. Core RNA polymerase was spotted onto nitrocellulose membranes and immunoblotted by using anti-TxeR antibodies after incubation with crude extract of E. coli (200 μg) carrying the vector pET28-b (filter 1), crude extract of E. coli carrying pCD54 (expressing TxeR) (filters 2–4), or heat-inactivated crude extract of TxeR-expressing cells (filter 5). The triangle indicates the use of increasing amounts of crude extract of_E. coli_ carrying pCD54 (50, 100, and 200 μg).
Figure 8
A model for toxin regulation in C. difficile. See text for details.
Similar articles
- Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors.
Dupuy B, Matamouros S. Dupuy B, et al. Res Microbiol. 2006 Apr;157(3):201-5. doi: 10.1016/j.resmic.2005.11.004. Epub 2005 Dec 29. Res Microbiol. 2006. PMID: 16439101 Review. - Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression.
Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. Mani N, et al. J Bacteriol. 2002 Nov;184(21):5971-8. doi: 10.1128/JB.184.21.5971-5978.2002. J Bacteriol. 2002. PMID: 12374831 Free PMC article. - Molecular analysis of the promoter region of the Clostridium difficile toxin B gene that is functional in Escherichia coli.
Song KP, Faust C. Song KP, et al. J Med Microbiol. 1998 Apr;47(4):309-16. doi: 10.1099/00222615-47-4-309. J Med Microbiol. 1998. PMID: 9568996 - Integration of metabolism and virulence in Clostridium difficile.
Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. Bouillaut L, et al. Res Microbiol. 2015 May;166(4):375-83. doi: 10.1016/j.resmic.2014.10.002. Epub 2014 Oct 15. Res Microbiol. 2015. PMID: 25445566 Free PMC article. Review.
Cited by
- FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence.
Bouillaut L, Ramarao N, Buisson C, Gilois N, Gohar M, Lereclus D, Nielsen-Leroux C. Bouillaut L, et al. Appl Environ Microbiol. 2005 Dec;71(12):8903-10. doi: 10.1128/AEM.71.12.8903-8910.2005. Appl Environ Microbiol. 2005. PMID: 16332888 Free PMC article. - Predictive regulatory and metabolic network models for systems analysis of Clostridioides difficile.
Arrieta-Ortiz ML, Immanuel SRC, Turkarslan S, Wu WJ, Girinathan BP, Worley JN, DiBenedetto N, Soutourina O, Peltier J, Dupuy B, Bry L, Baliga NS. Arrieta-Ortiz ML, et al. Cell Host Microbe. 2021 Nov 10;29(11):1709-1723.e5. doi: 10.1016/j.chom.2021.09.008. Epub 2021 Oct 11. Cell Host Microbe. 2021. PMID: 34637780 Free PMC article. - Clostridium difficile infection: An overview of the disease and its pathogenesis, epidemiology and interventions.
Viswanathan VK, Mallozzi MJ, Vedantam G. Viswanathan VK, et al. Gut Microbes. 2010 Jul;1(4):234-242. doi: 10.4161/gmic.1.4.12706. Epub 2010 Jun 16. Gut Microbes. 2010. PMID: 21327030 Free PMC article. - Overview of current detection methods and microRNA potential in Clostridioides difficile infection screening.
Bocchetti M, Ferraro MG, Melisi F, Grisolia P, Scrima M, Cossu AM, Yau TO. Bocchetti M, et al. World J Gastroenterol. 2023 Jun 14;29(22):3385-3399. doi: 10.3748/wjg.v29.i22.3385. World J Gastroenterol. 2023. PMID: 37389232 Free PMC article. Review. - Effect of tcdR Mutation on Sporulation in the Epidemic Clostridium difficile Strain R20291.
Girinathan BP, Monot M, Boyle D, McAllister KN, Sorg JA, Dupuy B, Govind R. Girinathan BP, et al. mSphere. 2017 Feb 15;2(1):e00383-16. doi: 10.1128/mSphere.00383-16. eCollection 2017 Jan-Feb. mSphere. 2017. PMID: 28217744 Free PMC article.
References
- Kelly C P, LaMont J T. Annu Rev Med. 1998;49:375–390. - PubMed
- Von Eichel-Streiber C, Boquet P, Sauerborn M, Thelestam M. Trends Microbiol. 1996;4:375–382. - PubMed
- Just I, Wilm M, Selzer J, Rex G, Von Eichel-Streiber C, Mann M, Aktories K. J Biol Chem. 1995;270:13932–13936. - PubMed
- Wilkins T D, Lyerly D M. Trends Microbiol. 1996;4:49–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases