Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions - PubMed (original) (raw)
Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions
J W Thornton. Proc Natl Acad Sci U S A. 2001.
Abstract
The evolution of novelty in tightly integrated biological systems, such as hormones and their receptors, seems to challenge the theory of natural selection: it has not been clear how a new function for any one part (such as a ligand) can be selected for unless the other members of the system (e.g., a receptor) are already present. Here I show-based on identification and phylogenetic analysis of steroid receptors in basal vertebrates and reconstruction of the sequences and functional attributes of ancestral proteins-that the first steroid receptor was an estrogen receptor, followed by a progesterone receptor. Genome mapping and phylogenetic analyses indicate that the full complement of mammalian steroid receptors evolved from these ancient receptors by two large-scale genome expansions, one before the advent of jawed vertebrates and one after. Specific regulation of physiological processes by androgens and corticoids are relatively recent innovations that emerged after these duplications. These findings support a model of ligand exploitation in which the terminal ligand in a biosynthetic pathway is the first for which a receptor evolves; selection for this hormone also selects for the synthesis of intermediates despite the absence of receptors, and duplicated receptors then evolve affinity for these substances. In this way, novel hormone-receptor pairs are created, and an integrated system of increasing complexity elaborated. This model suggests that ligands for some "orphan" receptors may be found among intermediates in the synthesis of ligands for phylogenetically related receptors.
Figures
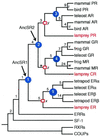
Figure 1
Phylogeny of the steroid receptor gene family. A reduced version is shown of the single most parsimonious phylogeny of 73 receptor sequences when the relative weight of gene duplications/losses to amino acids w > 3. (Length = 3,209 aa changes + 8 duplications + 0 losses. For unreduced phylogeny, see Fig. 7, which is published as supplemental data.) Support for each clade is shown as the number of extra steps required for the labeled node not to appear in the most parsimonious tree (19); all support values are insensitive to_w_ except *, shown for w = 10. Blue circles indicate gene duplications within the steroid receptor family; red squares mark the lamprey–gnathostome divergence; and unmarked nodes represent other speciation events. Ancestral steroid receptors are indicated. Italicized node labels correspond to Fig. 3. Tree length = 3,209 aa changes, eight duplications, zero losses; consistency index = 0.628; retention index = 0.870.
Figure 2
Steroid receptors diversified by large-scale genome expansions. Paralogous members of gene families with two or more members on the same chromosomes as the human 3-ketosteroid receptors are shown, without regard to map order.
Figure 3
Divergence rates of steroid receptor sequences after gene duplications indicate that the first steroid receptor was an estrogen receptor. Grouped bars show the ratio of the rate of amino acid replacement on the upper branch to that in the lower. White bars, rate ratio based on the relative rate test; black bars, ratio of parsimony branch lengths; hatched bars, ratio of maximum likelihood branch lengths. Outgroups for relative rate tests, from top to bottom, are estrogen-related receptors (ERRs), tetrapod ERs, lamprey PR, and lamprey GR. *, statistically significant departure from unity, P < 0.001. Parsimony and likelihood branch lengths are proportional to the number of weighted amino acid changes on paralogous branches that descend from duplication of an ancestral steroid receptor to an equivalent speciation event, with labels corresponding to nodes in Fig. 1.
Figure 4
Maximum likelihood reconstructions of ancestral sequences indicate that the first steroid receptor was an estrogen receptor and the first 3-ketosteroid receptor was not an androgen receptor. (a) Aligned amino acids forming the ligand-binding pocket of the ancestral steroid receptor (AncSR1), the ancestral 3-ketosteroid receptor (AncSR2), and five human steroid receptors, based on homology to human steroid receptors with solved structures. Colors and residue numbers refer to the positions shown in b. Filled circles (●) indicate amino acids identical to AncSR1; small dots are identical to AncSR2 but not AncSR1. Red, amino acids making direct hydrogen bonds with ligands (32, 33); blue, residues critical to discriminate androgens from C21 steroids in the androgen receptor (35). Amino acid numbers of homologous positions in the crystallized human receptors are at right; parentheses indicate positions that do not contact ligand in the indicated receptor. (b) Schematic of the ligand-binding pocket of ancestral steroid receptors with generic steroid hormone, based on homology to the crystal structures of human ERα and human PR (32, 33). Red and blue residues as in a. Yellow circles marked “R” indicate substituents that vary among steroid hormones.
Figure 5
Reconstruction of ligand-binding characteristics of ancestral steroid receptors indicates that the ancestral 3-ketosteroid receptor did not bind corticoids. Substituents at critical positions of the ligands that each vertebrate receptor binds were coded as characters (Right). Character states at ancestral nodes were reconstructed on the reduced phylogeny of steroid receptor paralogs with a parsimony-based algorithm, and the inferred structures of the ligands bound by each ancestral receptor are shown (Left); colored groups correspond to characters in the matrix. ?, substituent groups that could not be unambiguously reconstructed; NA, not applicable. P, progesterone; E2, estradiol; T, testosterone; B, corticosterone; F, cortisol; Aldo, aldosterone.
Similar articles
- Recent insights into the origins of adrenal and sex steroid receptors.
Baker ME. Baker ME. J Mol Endocrinol. 2002 Jun;28(3):149-52. doi: 10.1677/jme.0.0280149. J Mol Endocrinol. 2002. PMID: 12063181 Review. - Evolution of hormone-receptor complexity by molecular exploitation.
Bridgham JT, Carroll SM, Thornton JW. Bridgham JT, et al. Science. 2006 Apr 7;312(5770):97-101. doi: 10.1126/science.1123348. Science. 2006. PMID: 16601189 - Vestigialization of an allosteric switch: genetic and structural mechanisms for the evolution of constitutive activity in a steroid hormone receptor.
Bridgham JT, Keay J, Ortlund EA, Thornton JW. Bridgham JT, et al. PLoS Genet. 2014 Jan;10(1):e1004058. doi: 10.1371/journal.pgen.1004058. Epub 2014 Jan 9. PLoS Genet. 2014. PMID: 24415950 Free PMC article. - Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling.
Thornton JW, Need E, Crews D. Thornton JW, et al. Science. 2003 Sep 19;301(5640):1714-7. doi: 10.1126/science.1086185. Science. 2003. PMID: 14500980 - Steroid receptor phylogeny and vertebrate origins.
Baker ME. Baker ME. Mol Cell Endocrinol. 1997 Dec 12;135(2):101-7. doi: 10.1016/s0303-7207(97)00207-4. Mol Cell Endocrinol. 1997. PMID: 9484905 Review.
Cited by
- Diverse effects of phytoestrogens on the reproductive performance: cow as a model.
Wocławek-Potocka I, Mannelli C, Boruszewska D, Kowalczyk-Zieba I, Waśniewski T, Skarżyński DJ. Wocławek-Potocka I, et al. Int J Endocrinol. 2013;2013:650984. doi: 10.1155/2013/650984. Epub 2013 Apr 23. Int J Endocrinol. 2013. PMID: 23710176 Free PMC article. - Benefit:Risk Profile of Budesonide in Obstructive Airways Disease.
Tashkin DP, Lipworth B, Brattsand R. Tashkin DP, et al. Drugs. 2019 Nov;79(16):1757-1775. doi: 10.1007/s40265-019-01198-7. Drugs. 2019. PMID: 31549299 Free PMC article. Review. - Androgens, progestins, and glucocorticoids induce follicle-stimulating hormone beta-subunit gene expression at the level of the gonadotrope.
Thackray VG, McGillivray SM, Mellon PL. Thackray VG, et al. Mol Endocrinol. 2006 Sep;20(9):2062-79. doi: 10.1210/me.2005-0316. Epub 2006 May 4. Mol Endocrinol. 2006. PMID: 16675544 Free PMC article. - Hormone-activated estrogen receptors in annelid invertebrates: implications for evolution and endocrine disruption.
Keay J, Thornton JW. Keay J, et al. Endocrinology. 2009 Apr;150(4):1731-8. doi: 10.1210/en.2008-1338. Epub 2008 Nov 26. Endocrinology. 2009. PMID: 19036877 Free PMC article. - Histidyl-tRNA synthetase urzymes: Class I and II aminoacyl tRNA synthetase urzymes have comparable catalytic activities for cognate amino acid activation.
Li L, Weinreb V, Francklyn C, Carter CW Jr. Li L, et al. J Biol Chem. 2011 Mar 25;286(12):10387-95. doi: 10.1074/jbc.M110.198929. Epub 2011 Jan 26. J Biol Chem. 2011. PMID: 21270472 Free PMC article.
References
- Dawkins R. The Blind Watchmaker. New York: W. W. Norton; 1986.
- Kauffman S A. The Origins of Order: Self-Organization and Selection in Evolution. Oxford: Oxford Univ. Press; 1993.
- Thornton J W, DeSalle R. Syst Biol. 2000;49:183–201. - PubMed
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970.
- Fryxell K J. Trends Genet. 1996;12:364–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials