Regulatory functions of serine-46-phosphorylated HPr in Lactococcus lactis - PubMed (original) (raw)
Regulatory functions of serine-46-phosphorylated HPr in Lactococcus lactis
V Monedero et al. J Bacteriol. 2001 Jun.
Abstract
In most low-G+C gram-positive bacteria, the phosphoryl carrier protein HPr of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) becomes phosphorylated at Ser-46. This ATP-dependent reaction is catalyzed by the bifunctional HPr kinase/P-Ser-HPr phosphatase. We found that serine-phosphorylated HPr (P-Ser-HPr) of Lactococcus lactis participates not only in carbon catabolite repression of an operon encoding a beta-glucoside-specific EII and a 6-P-beta-glucosidase but also in inducer exclusion of the non-PTS carbohydrates maltose and ribose. In a wild-type strain, transport of these non-PTS carbohydrates is strongly inhibited by the presence of glucose, whereas in a ptsH1 mutant, in which Ser-46 of HPr is replaced with an alanine, glucose had lost its inhibitory effect. In vitro experiments carried out with L. lactis vesicles had suggested that P-Ser-HPr is also implicated in inducer expulsion of nonmetabolizable homologues of PTS sugars, such as methyl beta-D-thiogalactoside (TMG) and 2-deoxy-D-glucose (2-DG). In vivo experiments with the ptsH1 mutant established that P-Ser-HPr is not necessary for inducer expulsion. Glucose-activated 2-DG expulsion occurred at similar rates in wild-type and ptsH1 mutant strains, whereas TMG expulsion was slowed in the ptsH1 mutant. It therefore seems that P-Ser-HPr is not essential for inducer expulsion but that in certain cases it can play an indirect role in this regulatory process.
Figures
FIG. 1
Construction of an L. lactis strain expressing Ser46Ala mutant HPr. The wild-type strain MG5267 was transformed with plasmid pNZ9290 (26), which contains the 5′ part of the L. lactis ptsl gene and several hundred base pairs of the upstream region of the ptsHI operon. The ptsH gene located in front of _pts_I is partly deleted and the deleted region is replaced with an erythromycin resistance cassette. After a double-crossover recombination, a ptsH::erm strain (LlG100) was obtained. This strain exhibited a _pts_-negative phenotype and was transformed with the thermosensitive plasmid pGhostS46A, which carries the ptsH1 allele (the position of the Ser46Ala mutation is indicated with a triangle). After two successive recombination events, a pts+ strain carrying the ptsH1 allele (LlG101) was obtained.
FIG. 2
Western blot with L. lactis crude extracts prepared from glucose-grown wild-type and ptsH1 mutant strains and separated on a nondenaturing polyacrylamide gel. The various forms of HPr were detected with antibodies raised against B. subtilis HPr. Crude extracts from L. lactis wild-type MG5267 (lanes 1 and 2) and L. lactis ptsH1 mutant LlG101 (lanes 3 and 4) are shown. Extracts separated in lanes 2 and 4 were heated for 10 min at 65°C before they were loaded onto the gel.
FIG. 3
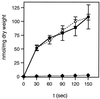
Transport of [14C]glucose (1 mM) by the L. lactis wild-type strain MG5267 (filled squares), the ptsH::erm disruption strain LlG100 (filled rhombs), and the ptsH1 mutant LlG101 (open circles). Cells were grown in M17 medium containing 0.5% glucose.
FIG. 4
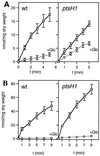
Transport of 14C-labeled TMG and mannitol and their exclusion by glucose in the L. lactis MG5267 (wild-type [wt]) and LlG101 (ptsH1 mutant) strains. (A) TMG transport with cells grown in M17 medium containing 0.5% lactose; (B) mannitol transport with cells grown in M17 medium containing 0.5% mannitol. Transport assays were carried out in the absence of glucose (squares) or with 10 mM glucose added 1 min prior to adding the radiolabeled sugar (circles). In panel B, the error bars for the experiments carried out in the presence of glucose were too small to be drawn by the program.
FIG. 5
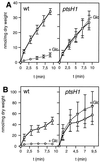
Transport of the 14C-labeled non-PTS sugars ribose (A) and maltose (B) and their exclusion by 10 mM glucose in L. lactis MG5267 (wild-type) and LlG101 (ptsH1 mutant) strains. Transport assays were carried out in the absence of glucose (squares) or with 10 mM glucose added 1 min prior to adding the radiolabeled sugar (circles). Cells were grown in M17 medium containing 0.5% ribose (A) or 0.5% maltose (B).
FIG. 6
Autoradiogram showing the amounts of [14C]TMG and [14C]TMG-6-P present in L. lactis cells and in the medium before and after inducer expulsion. [14C]TMG and [14C]TMG-6-P were separated by thin-layer chromatography. Lanes 1 and 3, [14C]TMG-6-P accumulated in wild-type and ptsH1 mutant cells; lanes 2 and 4, [14C]TMG present in cells and in the medium after 5 min of expulsion. Expulsion experiments were carried out with the wild-type strain MG5267 (lanes 1 and 2) and the ptsH1 mutant LlG101 (lanes 3 and 4). The cells were grown in 0.5% lactose-containing M17 medium.
FIG. 7
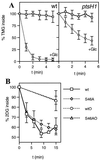
Expulsion of accumulated [14C]TMG-6-P (A) and [14C]2-DG-6-P (B) in the L. lactis wild-type strain MG5267 and the ptsH1 mutant LlG101. Cells grown in the presence of 0.5% lactose or 0.5% glucose were preloaded with [14C]TMG or [14C]2-DG, respectively. The amount of labeled sugar remaining inside the cells during a 5-min (for TMG) or 15-min (for 2-DG) incubation period at 37°C in the presence or absence of glucose was determined by withdrawing aliquots at the indicated time intervals and analyzing them by the rapid-filtration method (37). Squares, no sugar added; circles, 10 mM glucose added at time zero. S46AO and wtO, no glucose added to cells preloaded with [14C]2-DG-6-P. For the latter samples, aliquots were withdrawn only at the beginning and at the end of the experiments. Leakage levels of [14C]2-DG-6-P from the cells were found to be nearly identical for the wild-type and the ptsH1 mutant strains, explaining why the lines for the two strains coincide.
Similar articles
- Phosphorylation of HPr by the bifunctional HPr Kinase/P-ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion.
Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martínez G, Deutscher J. Dossonnet V, et al. J Bacteriol. 2000 May;182(9):2582-90. doi: 10.1128/JB.182.9.2582-2590.2000. J Bacteriol. 2000. PMID: 10762262 Free PMC article. - Catabolite repression and inducer control in Gram-positive bacteria.
Saier MH, Chauvaux S, Cook GM, Deutscher J, Paulsen IT, Reizer J, Ye JJ. Saier MH, et al. Microbiology (Reading). 1996 Feb;142 ( Pt 2):217-230. doi: 10.1099/13500872-142-2-217. Microbiology (Reading). 1996. PMID: 8932696 Review. - Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria.
Boël G, Mijakovic I, Mazé A, Poncet S, Taha MK, Larribe M, Darbon E, Khemiri A, Galinier A, Deutscher J. Boël G, et al. J Mol Microbiol Biotechnol. 2003;5(4):206-15. doi: 10.1159/000071072. J Mol Microbiol Biotechnol. 2003. PMID: 12867744 Review.
Cited by
- Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose.
Gopal S, Berg D, Hagen N, Schriefer EM, Stoll R, Goebel W, Kreft J. Gopal S, et al. PLoS One. 2010 Apr 27;5(4):e10349. doi: 10.1371/journal.pone.0010349. PLoS One. 2010. PMID: 20436965 Free PMC article. - Deciphering the Regulation of the Mannitol Operon Paves the Way for Efficient Production of Mannitol in Lactococcus lactis.
Xiao H, Bang-Berthelsen CH, Jensen PR, Solem C. Xiao H, et al. Appl Environ Microbiol. 2021 Jul 27;87(16):e0077921. doi: 10.1128/AEM.00779-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34105983 Free PMC article. - How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria.
Deutscher J, Francke C, Postma PW. Deutscher J, et al. Microbiol Mol Biol Rev. 2006 Dec;70(4):939-1031. doi: 10.1128/MMBR.00024-06. Microbiol Mol Biol Rev. 2006. PMID: 17158705 Free PMC article. Review. - GlaR (YugA)-a novel RpiR-family transcription activator of the Leloir pathway of galactose utilization in Lactococcus lactis IL1403.
Aleksandrzak-Piekarczyk T, Szatraj K, Kosiorek K. Aleksandrzak-Piekarczyk T, et al. Microbiologyopen. 2019 May;8(5):e00714. doi: 10.1002/mbo3.714. Epub 2018 Aug 11. Microbiologyopen. 2019. PMID: 30099846 Free PMC article. - Engineering Lactococcus lactis for production of mannitol: high yields from food-grade strains deficient in lactate dehydrogenase and the mannitol transport system.
Gaspar P, Neves AR, Ramos A, Gasson MJ, Shearman CA, Santos H. Gaspar P, et al. Appl Environ Microbiol. 2004 Mar;70(3):1466-74. doi: 10.1128/AEM.70.3.1466-1474.2004. Appl Environ Microbiol. 2004. PMID: 15006767 Free PMC article.
References
- Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases