Multiple maternal origins and weak phylogeographic structure in domestic goats - PubMed (original) (raw)
Multiple maternal origins and weak phylogeographic structure in domestic goats
G Luikart et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Domestic animals have played a key role in human history. Despite their importance, however, the origins of most domestic species remain poorly understood. We assessed the phylogenetic history and population structure of domestic goats by sequencing a hypervariable segment (481 bp) of the mtDNA control region from 406 goats representing 88 breeds distributed across the Old World. Phylogeographic analysis revealed three highly divergent goat lineages (estimated divergence >200,000 years ago), with one lineage occurring only in eastern and southern Asia. A remarkably similar pattern exists in cattle, sheep, and pigs. These results, combined with recent archaeological findings, suggest that goats and other farm animals have multiple maternal origins with a possible center of origin in Asia, as well as in the Fertile Crescent. The pattern of goat mtDNA diversity suggests that all three lineages have undergone population expansions, but that the expansion was relatively recent for two of the lineages (including the Asian lineage). Goat populations are surprisingly less genetically structured than cattle populations. In goats only approximately 10% of the mtDNA variation is partitioned among continents. In cattle the amount is >/=50%. This weak structuring suggests extensive intercontinental transportation of goats and has intriguing implications about the importance of goats in historical human migrations and commerce.
Figures
Figure 1
Neighbor-joining tree of mtDNA types from 406 domestic goats and 14 wild Capra. Trees constructed by using other methods (e.g., UPGMA or neighbor-joining with alpha = 0.20–0.40) were nearly identical in shape. The large star-shaped cluster (C. hircus A) contains 316 mtDNA types (found in 370 individuals and in all breeds). The two smaller lineages (C. hircus B and C) contain only eight and seven mtDNA types (found in 25 and 11 individuals, respectively). C. hircus B was detected only in eastern and southern Asia. C. hircus C was found in Mongolia, Switzerland, and Slovenia (Fig. 2). Numbers on branches are the percent of 2,000 bootstrap trees with the same branch structure. Only bootstrap values >70 are given. The wild taxon with sequences most similar to domestic goats is Capra aegagrus (61.3 substitutions, on average, using the gamma-corrected distance). The second most similar taxon is_Capra cylindricornis_ (84.5 substitutions; see Table 4, which is published as supplemental data on the PNAS web site,
). It is not surprising that some wild taxa appear to be paraphyletic because (i) the taxonomy of_Capra_ is very poorly understood and erroneous taxonomic classifications are possible (7), (ii) paraphyly has been reported (33, 34), and (iii) intertaxon hybridization is possible (23) and is thought to occur in Daghestan where our (paraphyletic) samples originated.
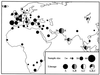
Figure 2
Geographic distribution of samples and of the three mtDNA lineages. The size of each circle is proportional to the sample size (1 to 62) from each of 44 countries. The presence of each lineage in a country is represented by a different color (black, lineage A; gray, lineage B; white, lineage C). Thus, the Asian B lineage occurs in Pakistan, India, Malaysia, and Mongolia. The numbers beside each circle on the map show the number of individuals from each lineage. The complete list of breeds and number of individuals sampled per breed and country are shown in Table 3, which is published a supplemental data.
Figure 3
Mismatch distributions (i.e., pairwise sequence-difference distributions) for mtDNA types from the major lineage of goat sequences, C. hircus A, and from the two smaller lineages, C. hircus B and C. A signature of population growth (i.e., a bell-shaped distribution) is clearly evident in the distribution for all three clusters of sequences, as would be expected for populations expanding after the domestication of relatively few founder-individuals (25). The means of the distributions are different, suggesting different expansion dates for each of the three goat lineages. The Asian lineage (C. hircus B) displays the most recent expansion date.
Comment in
- Livestock genetic origins: goats buck the trend.
MacHugh DE, Bradley DG. MacHugh DE, et al. Proc Natl Acad Sci U S A. 2001 May 8;98(10):5382-4. doi: 10.1073/pnas.111163198. Proc Natl Acad Sci U S A. 2001. PMID: 11344280 Free PMC article. No abstract available.
Similar articles
- Mitochondrial diversity and phylogeographic structure of Chinese domestic goats.
Chen SY, Su YH, Wu SF, Sha T, Zhang YP. Chen SY, et al. Mol Phylogenet Evol. 2005 Dec;37(3):804-14. doi: 10.1016/j.ympev.2005.06.014. Epub 2005 Jul 26. Mol Phylogenet Evol. 2005. PMID: 16051502 - Divergent mtDNA lineages of goats in an Early Neolithic site, far from the initial domestication areas.
Fernández H, Hughes S, Vigne JD, Helmer D, Hodgins G, Miquel C, Hänni C, Luikart G, Taberlet P. Fernández H, et al. Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15375-9. doi: 10.1073/pnas.0602753103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030824 Free PMC article. - Tracing the history of goat pastoralism: new clues from mitochondrial and Y chromosome DNA in North Africa.
Pereira F, Queirós S, Gusmão L, Nijman IJ, Cuppen E, Lenstra JA; Econogene Consortium; Davis SJ, Nejmeddine F, Amorim A. Pereira F, et al. Mol Biol Evol. 2009 Dec;26(12):2765-73. doi: 10.1093/molbev/msp200. Epub 2009 Sep 3. Mol Biol Evol. 2009. PMID: 19729424 - Conservation genetics of cattle, sheep, and goats.
Taberlet P, Coissac E, Pansu J, Pompanon F. Taberlet P, et al. C R Biol. 2011 Mar;334(3):247-54. doi: 10.1016/j.crvi.2010.12.007. Epub 2011 Feb 1. C R Biol. 2011. PMID: 21377620 Review. - The evolution of contemporary livestock species: Insights from mitochondrial genome.
Jain K, Panigrahi M, Nayak SS, Rajawat D, Sharma A, Sahoo SP, Bhushan B, Dutt T. Jain K, et al. Gene. 2024 Nov 15;927:148728. doi: 10.1016/j.gene.2024.148728. Epub 2024 Jun 27. Gene. 2024. PMID: 38944163 Review.
Cited by
- Mitochondrial DNA evidence indicates the local origin of domestic pigs in the upstream region of the Yangtze River.
Jin L, Zhang M, Ma J, Zhang J, Zhou C, Liu Y, Wang T, Jiang AA, Chen L, Wang J, Jiang Z, Zhu L, Shuai S, Li R, Li M, Li X. Jin L, et al. PLoS One. 2012;7(12):e51649. doi: 10.1371/journal.pone.0051649. Epub 2012 Dec 13. PLoS One. 2012. PMID: 23272130 Free PMC article. - Genetic analysis reveals the wild ancestors of the llama and the alpaca.
Kadwell M, Fernandez M, Stanley HF, Baldi R, Wheeler JC, Rosadio R, Bruford MW. Kadwell M, et al. Proc Biol Sci. 2001 Dec 22;268(1485):2575-84. doi: 10.1098/rspb.2001.1774. Proc Biol Sci. 2001. PMID: 11749713 Free PMC article. - Genetic Diversity and Population Structure of Llamas (Lama glama) from the Camelid Germplasm Bank-Quimsachata.
Paredes GF, Yalta-Macedo CE, Gutierrez GA, Veli-Rivera EA. Paredes GF, et al. Genes (Basel). 2020 May 12;11(5):541. doi: 10.3390/genes11050541. Genes (Basel). 2020. PMID: 32408471 Free PMC article. - Lymnaea schirazensis, an overlooked snail distorting fascioliasis data: genotype, phenotype, ecology, worldwide spread, susceptibility, applicability.
Bargues MD, Artigas P, Khoubbane M, Flores R, Glöer P, Rojas-García R, Ashrafi K, Falkner G, Mas-Coma S. Bargues MD, et al. PLoS One. 2011;6(9):e24567. doi: 10.1371/journal.pone.0024567. Epub 2011 Sep 29. PLoS One. 2011. PMID: 21980347 Free PMC article. - Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability.
Colli L, Lancioni H, Cardinali I, Olivieri A, Capodiferro MR, Pellecchia M, Rzepus M, Zamani W, Naderi S, Gandini F, Vahidi SM, Agha S, Randi E, Battaglia V, Sardina MT, Portolano B, Rezaei HR, Lymberakis P, Boyer F, Coissac E, Pompanon F, Taberlet P, Ajmone Marsan P, Achilli A. Colli L, et al. BMC Genomics. 2015 Dec 29;16:1115. doi: 10.1186/s12864-015-2342-2. BMC Genomics. 2015. PMID: 26714643 Free PMC article.
References
- Pringle H. Science. 1998;282:1448.
- Porter V. Goats of the World. Ipswich, U.K.: Farming Press; 1996.
- Vigne J-D, Buitenhuis H. Paléorient. 1999;25:49–62.
- Peters J, Helmer D, von den Driesch A, Sana-Segui M. Paléorient. 1999;25:27–47.
- Zeder M A, Hesse B. Science. 2000;287:2254–2257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources