Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion - PubMed (original) (raw)
Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion
A Becskei et al. EMBO J. 2001.
Abstract
Feedback is a ubiquitous control mechanism of gene networks. Here, we have used positive feedback to construct a synthetic eukaryotic gene switch in Saccharomyces cerevisiae. Within this system, a continuous gradient of constitutively expressed transcriptional activator is translated into a cell phenotype switch when the activator is expressed autocatalytically. This finding is consistent with a mathematical model whose analysis shows that continuous input parameters are converted into a bimodal probability distribution by positive feedback, and that this resembles analog-digital conversion. The autocatalytic switch is a robust property in eukaryotic gene expression. Although the behavior of individual cells within a population is random, the proportion of the cell population displaying either low or high expression states can be regulated. These results have implications for understanding the graded and probabilistic mechanisms of enhancer action and cell differentiation.
Figures
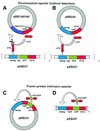
Fig. 1. Design of expression systems. (A) Constitutive system. (B, C and D) Autocatalytic systems. Blue colors represent promoter constructs (tetreg, CMV and CYC1), green for GFP, and red for the modules of activator. rtTA consists of a DNA binding domain, the reverse TetR (TetR) and an activator domain, VP16ad (VP16) (Gossen et al., 1995). The reporter construct trunc–GFP consists of an N-terminal tag, GFP and VP16ad. The CEN-TRP1 plasmids pBB140, pBB240, pBB340 and pBB247 contain the expression cassettes CMV-rtTA, tetreg-rtTA, CYC1-rtTA and tetreg-rtTA–GFP, respectively. The integrative LEU2 plasmids pAB237 and pAB247 are obtained by insertion of tetreg-trunc–GFP and tetreg-rtTA–GFP, respectively. Integration of pAB237 and pAB247 into the LEU2 locus of GFY259-2B resulted in yeast strains ABY001–ABY020 and ABY021–ABY040, respectively. Strains with the same number of integrated repeats (n) showed consistent behavior.
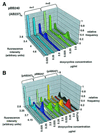
Fig. 2. Distribution of cell fluorescence intensities in chromosomal reporter systems. (A) The constitutive plasmid pBB140 (CMV promoter) was transformed into strains ABY001 (one copy of the reporter construct, pAB237) (blue shades) and ABY016 (more than five copies of reporter construct, pAB237) (yellow shades). Doxycycline was added to the culture to yield a final concentration of 0.025, 0.25 and 2.5 µg/ml. The distributions at 0.025 and zero doxycycline concentrations are very similar, which might be explained by the residual binding of rtTA to the enhancer in the absence of inducer. Cultures were grown in SD-Leu, Trp. Relative cell count was obtained by dividing the actual frequency by the frequency at the mode of distribution. In this way the differences in the mean intensities are visualized better. The mean value (m) and standard deviation (s) for ABY016 at 2.5 µg/ml inducer concentration are m = 4.55 and s = 0.159. The standard deviation indicates the width of distribution. (B) Comparison of the autocatalytic (pBB240) and constitutive (pBB340) systems transformed into the ABY016 strain. Both expression systems contain the CYC1 core promoter. The autocatalytic and constitutive systems are labeled with ‘autocat’ and ‘constit’ next to the indicated inducer concentration. For pBB340, m = 3.67 and s = 0.240 at 2.5 µg/ml inducer concentration.
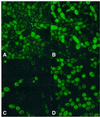
Fig. 3. Fluorescence microscope image of cells. Panels were obtained by merging phase contrast and fluorescence images of ABY016 cells. (A and B) Cells contain pBB140 and are induced with 0.25 (A) and 2.5 µg/ml (B) doxycycline, and exposed for 25 ms. (C and D) Cells contain pBB240 and are induced with 0.25 (C) and 2.5 µg/ml (D) doxycycline, and exposed for 10 ms.
Fig. 4. Fluorescence level distributions in the autocatalytic systems. (A) Chromosomal reporter system. pBB240 was transformed into strains ABY001 (one copy of the reporter gene) and ABY011 (two copies of the reporter gene), and are represented by blue and green shades, respectively. Cells were grown in SD-Leu, Trp. (B) Intrinsic reporter system. Out of the series ABY021–040, ABY021 and ABY022 were examined with one and more than five copies of pAB247, and are represented by blue and yellow shades, respectively. Strains were grown in SD-Leu. pBB247 was transformed into GFY259-2B, grown in SD-Trp; results are represented by green shades. The black lane represents the galactose-regulated plasmid pBB407 (see Materials and methods).
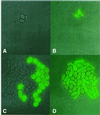
Fig. 5. Fate of single cells during growth. The division of non-fluorescent and fluorescent ABY001 cells was followed on agarose layers containing 2.5 µg/ml doxycycline. (A and B) Image of cells at the start of the experiment. (C) Colony derived from cells on (A) after 16 h of incubation. (D) Colony derived from cells on (B) after 22 h of incubation.
Fig. 6. Approximate probability distribution in an autocatalytic system. The approximation is based on the negatively signed potential V(x) function. Regions of attraction of the lower and upper steady-states are indicated. Values of V(x) <0 are shown. _V_(_x_) is obtained from ϕ(_x_) [equation (2), Materials and methods] by normalization by the sum of the lowest values of the two potential wells, which correspond to the lower and upper steady-states. The parameter values are _d_ = 10, _r_ = 0.1 min–1, _k_ = 1 min–1. To simulate different degrees of activation, _s_ took values of 5, 10 and 20 min–1. Concentrations and concentration-based parameters are non-dimensional. The range of bistability is 4 < _s_ < 148 min–1, which encompasses almost two orders of magnitude of protein production rate. Outside this region, a single stable state exists as a continuation of the lower and upper steady-states at _s_ <4 and _s_ >148 min–1, respectively. The approximate values of the steady-states are 100.67, 100.9 and 101.3 at s = 5, 10 and 20 min–1, respectively, indicating the rise of the upper steady-state parallel with the activation level.
Similar articles
- Cell signaling can direct either binary or graded transcriptional responses.
Biggar SR, Crabtree GR. Biggar SR, et al. EMBO J. 2001 Jun 15;20(12):3167-76. doi: 10.1093/emboj/20.12.3167. EMBO J. 2001. PMID: 11406593 Free PMC article. - Graded mode of transcriptional induction in yeast pheromone signalling revealed by single-cell analysis.
Poritz MA, Malmstrom S, Kim MK, Rossmeissl PJ, Kamb A. Poritz MA, et al. Yeast. 2001 Oct;18(14):1331-8. doi: 10.1002/yea.777. Yeast. 2001. PMID: 11571757 - Why Ppr1p is a weak activator of transcription.
Pätzold AJ, Lehming N. Pätzold AJ, et al. FEBS Lett. 2001 Apr 6;494(1-2):64-8. doi: 10.1016/s0014-5793(01)02312-2. FEBS Lett. 2001. PMID: 11297736 - Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae).
Chambers A, Packham EA, Graham IR. Chambers A, et al. Curr Genet. 1995 Dec;29(1):1-9. doi: 10.1007/BF00313187. Curr Genet. 1995. PMID: 8595651 Review. No abstract available. - Gene identification using the yeast two-hybrid system.
Bai C, Elledge SJ. Bai C, et al. Methods Enzymol. 1996;273:331-47. doi: 10.1016/s0076-6879(96)73029-x. Methods Enzymol. 1996. PMID: 8791622 Review. No abstract available.
Cited by
- Dynamics of transcription driven by the tetA promoter, one event at a time, in live Escherichia coli cells.
Muthukrishnan AB, Kandhavelu M, Lloyd-Price J, Kudasov F, Chowdhury S, Yli-Harja O, Ribeiro AS. Muthukrishnan AB, et al. Nucleic Acids Res. 2012 Sep 1;40(17):8472-83. doi: 10.1093/nar/gks583. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730294 Free PMC article. - A computational model of stem cells' internal mechanism to recapitulate spatial patterning and maintain the self-organized pattern in the homeostasis state.
Khorasani N, Sadeghi M. Khorasani N, et al. Sci Rep. 2024 Jan 17;14(1):1528. doi: 10.1038/s41598-024-51386-z. Sci Rep. 2024. PMID: 38233402 Free PMC article. - An optimal number of molecules for signal amplification and discrimination in a chemical cascade.
Morishita Y, Kobayashi TJ, Aihara K. Morishita Y, et al. Biophys J. 2006 Sep 15;91(6):2072-81. doi: 10.1529/biophysj.105.070797. Epub 2006 Jun 23. Biophys J. 2006. PMID: 16798800 Free PMC article. - Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems.
Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Bashor CJ, et al. Annu Rev Biophys. 2010;39:515-37. doi: 10.1146/annurev.biophys.050708.133652. Annu Rev Biophys. 2010. PMID: 20192780 Free PMC article. Review. - Improvement of the memory function of a mutual repression network in a stochastic environment by negative autoregulation.
Hasan ABMSU, Kurata H, Pechmann S. Hasan ABMSU, et al. BMC Bioinformatics. 2019 Dec 27;20(1):734. doi: 10.1186/s12859-019-3315-2. BMC Bioinformatics. 2019. PMID: 31881978 Free PMC article.
References
- A-Mohammadi S. and Hawkins,R.E. (1998) Efficient transgene regulation from a single tetracycline-controlled positive feedback regulatory system. Gene Ther., 5, 76–84. - PubMed
- Bateman E. (1998) Autoregulation of eukaryotic transcription factors. Prog. Nucleic Acid Res. Mol. Biol., 60, 133–168. - PubMed
- Becskei A. and Serrano,L. (2000) Engineering stability in gene networks by autoregulation. Nature, 405, 590–593. - PubMed
- Bouhassira E.E., Westerman,K. and Leboulch,P. (1997) Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood, 90, 3332–3344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases