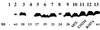Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8 - PubMed (original) (raw)
Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8
R B Kumar et al. J Bacteriol. 2001 Jun.
Abstract
The VirB8 protein of Agrobacterium tumefaciens is essential for DNA transfer to plants. VirB8, a 237-residue polypeptide, is an integral membrane protein with a short N-terminal cytoplasmic domain. It interacts with two transport pore proteins, VirB9 and VirB10, in addition to itself. To study the role of these interactions in DNA transfer and to identify essential amino acids of VirB8, we introduced random mutations in virB8 by the mutagenic PCR method. The putative mutants were tested for VirB8 function by the ability to complement a virB8 deletion mutant in tumor formation assays. After multiple rounds of screening 13 mutants that failed to complement the virB8 deletion mutation were identified. Analysis of the mutant strains by DNA sequence analysis, Western blot assays, and reconstruction of new point mutations led to the identification of five amino acid residues that are essential for VirB8 function. The substitution of glycine-78 to serine, serine-87 to leucine, alanine-100 to valine, arginine-107 to proline or alanine, and threonine-192 to methionine led to the loss of VirB8 activity. When introduced into the wild-type strain, virB8(S87L) partially suppressed the tumor forming ability of the wild-type protein. Analysis of protein-protein interaction by the yeast two-hybrid assay indicated that VirB8(R107P) is defective in interactions with both VirB9 and VirB10. A second mutant VirB8(S87L) is defective in interaction with VirB9.
Figures
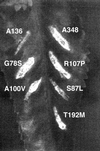
FIG. 1
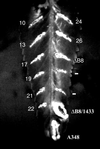
Phenotype of the virB8 mutants. The virB8 mutants were tested for the ability to complement a deletion in virB8. The mutants in A. tumefaciens PC1008 (ΔB8) were used to infect K. daigremontiana leaves and scored for tumor formation 3 weeks after infection. A subset of the mutants listed on Table 1 is shown. The numbers indicate the mutant number. Plasmid pAD1433 or its derivative harbors virB8 or the mutant. A348, wild-type strain; −, uninfected wound site.
FIG. 2
Effect of the virB8 mutations on protein stability. The level of VirB8 and its mutants were monitored by Western blot assays using purified VirB8 antibodies (16). Lanes 1, 2, and 13, uninduced A348, induced A348, and PC1008/pAD1433, respectively; lanes 3 to 12, virB8 mutants 10, 13, 17, 19, 21, 24, 26, _virB8_S87L, _virB8_T192M, and _virB8_R107A, respectively.
FIG. 3
Role of VirB8 arginine-107 in DNA transfer to plants. An arginine at position 107 of VirB8 was changed to alanine by site-specific mutagenesis (17). The mutants were tested by complementation assays. wt, wild-type virB8; R107A, _virB8_R107A.
FIG. 4
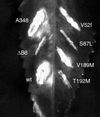
Identification of amino acids responsible for the avirulent phenotype of the double mutants. Mutations that led to single-amino-acid substitutions in virB8 were introduced by site-specific mutagenesis, and the mutants were introduced into A. tumefaciens PC1008. The resultant strains were used to infect K. daigremontiana leaves. The strains used for infection were A. tumefaciens A348 (A348), PC1008 (ΔB8), and PC1008 harboring a plasmid that expresses wild-type virB8 (wt), _virB8_V52I (V52I), _virB8_S87L (S87L), _virB8_V189M (V189M), or _virB_T192M (T192M).
FIG. 5
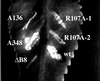
Dominant-recessive phenotype of the virB8 mutants. A plasmid expressing virB8 or its mutant was introduced into A. tumefaciens A348, and the resultant strains were used to infect K. daigremontiana leaves.
FIG. 6
Interaction of the VirB8 mutants with VirB8, VirB9, and VirB10. The interaction of VirB8 (wt) and its mutant with VirB8 (B8), VirB9 (B9), and VirB10 (B10) was monitored by the two-hybrid assay in yeast as described previously (10). A blue colony color indicates a positive interaction.
FIG. 7
Conservation of glycine-78 and sequences around arginine-107 in the VirB8 homologs. The amino acid sequence of a segment of A. tumefaciens VirB8 and its homologs is shown. Residues identical to the A. tumefaciens VirB8 sequence are shown as dots. Sequences that exhibited a high degree of conservation are boxed. Glycine-78, alanine-100, and arginine-107 are shown in boldface. The numbers on the left indicate the position of the first amino acid residue shown in the figure. The gaps were introduced to achieve maximum homology.
Similar articles
- Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration.
Mysore KS, Bassuner B, Deng XB, Darbinian NS, Motchoulski A, Ream W, Gelvin SB. Mysore KS, et al. Mol Plant Microbe Interact. 1998 Jul;11(7):668-83. doi: 10.1094/MPMI.1998.11.7.668. Mol Plant Microbe Interact. 1998. PMID: 9650299 - The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another.
Das A, Xie YH. Das A, et al. J Bacteriol. 2000 Feb;182(3):758-63. doi: 10.1128/JB.182.3.758-763.2000. J Bacteriol. 2000. PMID: 10633111 Free PMC article. - A single amino acid change in the transmembrane domain of the VirB8 protein affects dimerization, interaction with VirB10 and Brucella suis virulence.
Andrieux L, Bourg G, Pirone A, O'Callaghan D, Patey G. Andrieux L, et al. FEBS Lett. 2011 Aug 4;585(15):2431-6. doi: 10.1016/j.febslet.2011.07.004. Epub 2011 Jul 13. FEBS Lett. 2011. PMID: 21763312 - VirB8: a conserved type IV secretion system assembly factor and drug target.
Baron C. Baron C. Biochem Cell Biol. 2006 Dec;84(6):890-9. doi: 10.1139/o06-148. Biochem Cell Biol. 2006. PMID: 17215876 Review. - PCR systems for Agrobacterium tumefaciens detection.
Sachadyn P, Kur J. Sachadyn P, et al. Acta Microbiol Pol. 1997;46(2):129-43. Acta Microbiol Pol. 1997. PMID: 9429287 Review. No abstract available.
Cited by
- Structural Analysis and Inhibition of TraE from the pKM101 Type IV Secretion System.
Casu B, Smart J, Hancock MA, Smith M, Sygusch J, Baron C. Casu B, et al. J Biol Chem. 2016 Nov 4;291(45):23817-23829. doi: 10.1074/jbc.M116.753327. Epub 2016 Sep 15. J Biol Chem. 2016. PMID: 27634044 Free PMC article. - Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems.
Christie PJ. Christie PJ. Biochim Biophys Acta. 2004 Nov 11;1694(1-3):219-34. doi: 10.1016/j.bbamcr.2004.02.013. Biochim Biophys Acta. 2004. PMID: 15546668 Free PMC article. Review. - Biological diversity of prokaryotic type IV secretion systems.
Alvarez-Martinez CE, Christie PJ. Alvarez-Martinez CE, et al. Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review. - An anomalous type IV secretion system in Rickettsia is evolutionarily conserved.
Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. Gillespie JJ, et al. PLoS One. 2009;4(3):e4833. doi: 10.1371/journal.pone.0004833. Epub 2009 Mar 12. PLoS One. 2009. PMID: 19279686 Free PMC article. - Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori.
Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Terradot L, et al. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4596-601. doi: 10.1073/pnas.0408927102. Epub 2005 Mar 11. Proc Natl Acad Sci U S A. 2005. PMID: 15764702 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources