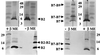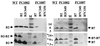VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus - PubMed (original) (raw)
VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus
V Sagulenko et al. J Bacteriol. 2001 Jun.
Abstract
Agrobacterium tumefaciens transfers oncogenic T-DNA and effector proteins to plant cells via a type IV secretion pathway. This transfer system, assembled from the products of the virB operon, is thought to consist of a transenvelope mating channel and the T pilus. When screened for the presence of VirB and VirE proteins, material sheared from the cell surface of octopine strain A348 was seen to possess detectable levels of VirB2 pilin, VirB5, and the VirB7 outer membrane lipoprotein. Material sheared from the cell surface of most virB gene deletion mutants also possessed VirB7, but not VirB2 or VirB5. During purification of the T pilus from wild-type cells, VirB2, VirB5, and VirB7 cofractionated through successive steps of gel filtration chromatography and sucrose density gradient centrifugation. A complex containing VirB2 and VirB7 was precipitated from a gel filtration fraction enriched for T pilus with both anti-VirB2 and anti-VirB7 antiserum. Both the exocellular and cellular forms of VirB7 migrated as disulfide-cross-linked dimers and monomers when samples were electrophoresed under nonreducing conditions. A mutant synthesizing VirB7 with a Ser substitution of the lipid-modified Cys15 residue failed to elaborate the T pilus, whereas a mutant synthesizing VirB7 with a Ser substitution for the disulfide-reactive Cys24 residue produced very low levels of T pilus. Together, these findings establish that the VirB7 lipoprotein localizes exocellularly, it associates with the T pilus, and both VirB7 lipid modification and disulfide cross-linking are important for T-pilus assembly. T-pilus-associated VirB2 migrated in nonreducing gels as a monomer and a disulfide-cross-linked homodimer, whereas cellular VirB2 migrated as a monomer. A strain synthesizing a VirB2 mutant with a Ser substitution for the reactive Cys64 residue elaborated T pilus but exhibited an attenuated virulence phenotype. Dithiothreitol-treated T pilus composed of native VirB2 pilin and untreated T pilus composed of the VirB2C64S mutant pilin distributed in sucrose gradients more predominantly in regions of lower sucrose density than untreated, native T pili. These findings indicate that intermolecular cross-linking of pilin monomers is not required for T-pilus production, but cross-linking does contribute to T-pilus stabilization.
Figures
FIG. 1
Identification of VirB2, VirB5, and VirB7 in the exocellular fraction obtained by shearing of A348 cells. Blots were developed with antisera to the VirB proteins listed, and an arrowhead marks the position of the corresponding VirB protein. Abbreviations: E, exocellular fraction containing surface organelles and proteins; C, cell pellet recovered after removal of surface material by shearing as described in the text; M, molecular mass markers, with sizes in kilodaltons indicated at the left.
FIG. 2
Purification of T pilus through sucessive steps of gel filtration chromatography and sucrose density gradient centrifugation. (A) Gel filtration fractions containing the VirB2, VirB5, and VirB7 proteins; the bottom blot is skewed slightly to the right relative to the top blot. (B) Sucrose density gradient fractions containing VirB2, VirB5, and VirB7. Top blots in each panel were developed with anti-VirB2 antiserum, and bottom blots were developed with anti-VirB5 and anti-VirB7 antisera. Positions of VirB proteins and sizes (in kilodaltons) of molecular mass markers (M) are denoted. Lane GF, gel filtration fraction 21 loaded onto the sucrose gradient.
FIG. 3
Transmission electron microscopy showing purified T pili obtained from sucrose density gradients. The first two images were from fraction 6 and second two were from fraction 8 of the sucrose gradient shown in Fig. 2. Arrowheads denote morphological features of interest, including single T pili (diameter, ∼10 nm), clumps of T pili, and occasional terminal sacculi as observed previously (20, 27). Pili were examined at a magnification of 40,000. Bar, 100 nm.
FIG. 4
Coprecipitation of a VirB2 and VirB7 complex presumptively corresponding to the T pilus with anti-VirB2 and anti-VirB5 antiserum from gel filtration fraction 21 (GF21) (see Fig. 2). Blots showing VirB2 and VirB7 proteins in complexes precipitated with anti-VirB2 antiserum (A) and with anti-VirB7 antiserum (B) are shown. Positions of VirB proteins and sizes (in kilodaltons) of molecular mass markers (M) are denoted. Abbreviations: S, supernatant fraction following precipitation; P, pellet recovered upon precipitation with the reagents listed at the top of each panel.
FIG. 5
Presence of the VirB7 homodimer in the exocellular fractions from the nonpolar Δ_virB_ mutants of A348 (4). Exocellular fractions were from wild-type strain A348 (lane WT) and PC1001 (Δ_virB1_) through PC1011 (Δ_virB11_) (lanes 1 through 11, respectively). Blots were developed with antisera to the VirB proteins denoted at the right. Samples analyzed with anti-VirB2 and anti-VirB5 antisera were electrophoresed under reducing conditions; samples analyzed with anti-VirB7 antisera were electrophoresed under nonreducing conditions to show the relative abundances of the VirB7 homodimer and monomer.
FIG. 6
Association of VirB2 and VirB7 homodimers with the T pilus. Top panels, VirB2 and VirB7 species present in extracts from A348 cells obtained after shearing to remove surface proteins and organelles. Extracts were electrophoresed under reducing (+βME) and nonreducing (−βME) conditions. The cellular form of VirB2 pilin migrates as a monomer, whereas the cellular form of VirB7 migrates predominantly as a disulfide-cross-linked dimer. A cross-reactive species of unknown composition is detected at a position corresponding to ∼18 kDa. Bottom panels, VirB2 and VirB7 species associated with T pili enriched from the exocellular fraction of A348 cells. The T-pilus-associated forms of both VirB2 and VirB7 migrated in nonreducing gels at positions corresponding to monomeric and disulfide-cross-linked homodimeric species. Positions of VirB2 and VirB7 species and sizes (in kilodaltons) of molecular mass markers are denoted.
FIG. 7
Effects of Cys-to-Ser substitution mutations on accumulation of exocellular VirB2 and VirB7. Exocellular fractions were from A348 (WT), strains PC1002 carrying pBB8 (B2) or pVS10 (B2C64S), and strains PC1007 carrying pPC974 (B7), pXZ14 (B7C24S), or pXZ16 (B7C15S). Top panels, protein samples were electrophoresed under reducing conditions. Bottom panels, protein samples were electrophoresed under nonreducing conditions. Blots on the left were developed with anti-VirB2 antiserum, and blots on the right were developed with anti-VirB7 antiserum. Sizes (in kilodaltons) of molecular mass markers (M) are denoted in the center.
FIG. 8
Effects of a Cys64Ser substitution mutation and DTT treatment of wild-type pilin on T-pilus assembly. (A) Virulence assays of PC1002 cells (Δ2) expressing wild-type VirB2 or VirB2C64S by inoculation of equivalent numbers of cells onto wounded Kalanchoe leaves. Cells synthesizing the C64S mutant incited tumors of variable sizes that were reproducibly smaller than those incited by cells synthesizing wild-type pilin. (B) T pilus composed of wild-type pilin enriched by gel filtration fraction chromatography was treated with 5 mM DTT, and both untreated and DTT-treated T-pilus samples were fractionated through identically prepared sucrose density gradients. (C) T pilus composed of wild-type or C64S mutant pilin present in the concentrated exocellular fractions from PC1002(pBB8) or PC1002(pVS10) cells, respectively, was fractionated through sucrose density gradients.
Similar articles
- Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion.
Jakubowski SJ, Krishnamoorthy V, Christie PJ. Jakubowski SJ, et al. J Bacteriol. 2003 May;185(9):2867-78. doi: 10.1128/JB.185.9.2867-2878.2003. J Bacteriol. 2003. PMID: 12700266 Free PMC article. - Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens.
Lai EM, Kado CI. Lai EM, et al. J Bacteriol. 1998 May;180(10):2711-7. doi: 10.1128/JB.180.10.2711-2717.1998. J Bacteriol. 1998. PMID: 9573157 Free PMC article. - Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens.
Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski PC, Baron C. Schmidt-Eisenlohr H, et al. J Bacteriol. 1999 Dec;181(24):7485-92. doi: 10.1128/JB.181.24.7485-7492.1999. J Bacteriol. 1999. PMID: 10601205 Free PMC article. - Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells.
Zupan JR, Ward D, Zambryski P. Zupan JR, et al. Curr Opin Microbiol. 1998 Dec;1(6):649-55. doi: 10.1016/s1369-5274(98)80110-0. Curr Opin Microbiol. 1998. PMID: 10066547 Review. - Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis.
Kado CI. Kado CI. Mol Microbiol. 1994 Apr;12(1):17-22. doi: 10.1111/j.1365-2958.1994.tb00990.x. Mol Microbiol. 1994. PMID: 7914664 Review.
Cited by
- Insights into the transcriptomic response of the plant engineering bacterium Ensifer adhaerens OV14 during transformation.
Zuniga-Soto E, Fitzpatrick DA, Doohan FM, Mullins E. Zuniga-Soto E, et al. Sci Rep. 2019 Jul 17;9(1):10344. doi: 10.1038/s41598-019-44648-8. Sci Rep. 2019. PMID: 31316079 Free PMC article. - Pathways of DNA Transfer to Plants from Agrobacterium tumefaciens and Related Bacterial Species.
Lacroix B, Citovsky V. Lacroix B, et al. Annu Rev Phytopathol. 2019 Aug 25;57:231-251. doi: 10.1146/annurev-phyto-082718-100101. Epub 2019 Jun 21. Annu Rev Phytopathol. 2019. PMID: 31226020 Free PMC article. - Transcriptional Activation of Virulence Genes of Rhizobium etli.
Wang L, Lacroix B, Guo J, Citovsky V. Wang L, et al. J Bacteriol. 2017 Feb 28;199(6):e00841-16. doi: 10.1128/JB.00841-16. Print 2017 Mar 15. J Bacteriol. 2017. PMID: 28069822 Free PMC article. - Potential of known and short prokaryotic protein motifs as a basis for novel peptide-based antibacterial therapeutics: a computational survey.
Ruhanen H, Hurley D, Ghosh A, O'Brien KT, Johnston CR, Shields DC. Ruhanen H, et al. Front Microbiol. 2014 Jan 21;5:4. doi: 10.3389/fmicb.2014.00004. eCollection 2014. Front Microbiol. 2014. PMID: 24478765 Free PMC article. - The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation.
Lacroix B, Citovsky V. Lacroix B, et al. Int J Dev Biol. 2013;57(6-8):467-81. doi: 10.1387/ijdb.130199bl. Int J Dev Biol. 2013. PMID: 24166430 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases