The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes - PubMed (original) (raw)
Comparative Study
. 2001 Jun;11(6):1018-33.
doi: 10.1101/gr.180401.
Affiliations
- PMID: 11381029
- PMCID: PMC311111
- DOI: 10.1101/gr.180401
Comparative Study
The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes
K Inoue et al. Genome Res. 2001 Jun.
Abstract
Duplication and deletion of the 1.4-Mb region in 17p12 that is delimited by two 24-kb low copy number repeats (CMT1A-REPs) represent frequent genomic rearrangements resulting in two common inherited peripheral neuropathies, Charcot-Marie-Tooth disease type 1A (CMT1A) and hereditary neuropathy with liability to pressure palsy (HNPP). CMT1A and HNPP exemplify a paradigm for genomic disorders wherein unique genome architectural features result in susceptibility to DNA rearrangements that cause disease. A gene within the 1.4-Mb region, PMP22, is responsible for these disorders through a gene-dosage effect in the heterozygous duplication or deletion. However, the genomic structure of the 1.4-Mb region, including other genes contained within the rearranged genomic segment, remains essentially uncharacterized. To delineate genomic structural features, investigate higher-order genomic architecture, and identify genes in this region, we constructed PAC and BAC contigs and determined the complete nucleotide sequence. This CMT1A/HNPP genomic segment contains 1,421,129 bp of DNA. A low copy number repeat (LCR) was identified, with one copy inside and two copies outside of the 1.4-Mb region. Comparison between physical and genetic maps revealed a striking difference in recombination rates between the sexes with a lower recombination frequency in males (0.67 cM/Mb) versus females (5.5 cM/Mb). Hypothetically, this low recombination frequency in males may enable a chromosomal misalignment at proximal and distal CMT1A-REPs and promote unequal crossing over, which occurs 10 times more frequently in male meiosis. In addition to three previously described genes, five new genes (TEKT3, HS3ST3B1, NPD008/CGI-148, CDRT1, and CDRT15) and 13 predicted genes were identified. Most of these predicted genes are expressed only in embryonic stages. Analyses of the genomic region adjacent to proximal CMT1A-REP indicated an evolutionary mechanism for the formation of proximal CMT1A-REP and the creation of novel genes by DNA rearrangement during primate speciation.
Figures
Figure 1
The genomic sequence map of the CMT1A duplication/HNPP deletion region in 17p12. The top solid horizontal line represents the genomic sequence of the CMT1A/HNPP region in the centromere to telomere orientation. Position 0 is assigned to the first base of the proximal CMT1A–REP and vertical markings are placed every 100 kb for reference. The STR polymorphic genetic markers are shown above. Shaded horizontal boxes below depict the large insert clones used to derive genomic sequences. Clones are identified by their individual names and GenBank accession numbers. Proximal and distal CMT1A–REPs are shown as vertical blue boxes, and newly identified low copy repeats (LCRA1, LCRA2, LCRB) are represented as vertical red bars. Arrowheads indicate the orientation/direction of each repeat unit. Underneath are shown known genes (green), predicted genes (purple), and pseudogenes (black), with arrows pointing in the direction of transcription. (MITE and HSMAR2-PMP22) mariner transposon-like elements; (CYPAP) cyclophilin A pseudogene; (60SRPL9P) 60S ribosomal protein L9 pseudogene; (60SRPL23AP) 60S ribosomal protein L23A pseudogene; (40SRPS18P) 40S ribosomal protein S18 pseudogene.
Figure 2
Genomic structure of LCRA1, LCRA2, and LCRB. (A) The genomic architecture of three LCRs is shown. High copy number retrotransposable elements including Alu, L1, MaLR, and MER2 type DNA elements, which are conserved between these LCR, are boxed in gray. Each exon of CDRT15 is shown as a solid black box. A small genomic region in the 5′ upstream of CDRT15 exon I representing DNA rearrangements (enlarged) between the three LCRs. The LCRA1 has a 132-bp region that is further divided into 25 bp (blue), 89 bp (pink) and 18 bp (green) segments. There are short tandem repeat sequences, 14 bp (orange) and 9 bp (red), flanking these subdivided fragments. In comparison with the same region in the LCRA2, the 25-bp monomer is tandemly duplicated and the 18-bp fragment is deleted in LCRA2. Furthermore, the distal boundary of 4.4-kb LCRB is located in this short genomic region between 14- and 9-bp repeat (arrow). These 18 bp are present in LCRB. (B) Hypothetical inversion involving LCRA1 and LCRA2 results in flipping of distal CMT1A–REP. The proximal and distal CMT1A–REPs are depicted by thick black arrows with their relative orientation given by the directions of arrows. LCRA1 and LCRA2 are depicted as open arrows. The genome architecture as determined by sequencing this specific genomic library alone placed the CMT1A–REPs in a direct orientation and thus the region in between is susceptible to duplication/deletion. Below is shown the hypothetical orientation of CMT1A–REPs if an inversion occurs via homologous recombination using LCRA1 and LCRA2 as substrates. Note that this orientation of CMT1A–REPs will prevent the formation of duplication/deletion event (Lupski 1998b).
Figure 3
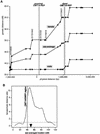
Sex-specific recombination frequencies in the CMT1A/HNPP genomic region. (A) The relationship between genetic and physical distance. The STR markers in the 17p12 CMT1A/HNPP region from the Marshfield genetic map were aligned to the nucleotide sequence-based physical map. The marker order is as follows: centromere-_D17S1794_-_D17S620_-_D17S2196_-_D17S1857_-_D17S953_-D17S1843
-_D17S793_-_D17S122_-_D17S918_-_D17S839_-_D17S955_-_D17S921_-D17S900
-_D17S922_-_D17S1856_-_D17S947_-_D17S936_-_D17S639_-_D17S799_-_D17S1808_-_D17S1803_-telomere (markers within the CMT1A/HNPP genomic region are underlined). Both CMT1A–REPs are shown as hatched bars. As the sequencing of the centromeric side of the map has not yet been completed, the physical distance was calculated on the basis of the BAC contig map (K. Inoue, K. Dewar, N. Katsanis, L.T. Reiter, E.S. Lander, K.L. Devon, D.W. Wyman, J.R. Lupski, and B. Birren, unpubl.). (B) Female/male distance ratio (vertical axis) is plotted along the sex-averaged genetic map (horizontal axis). The histogram was obtained from the Marshfield Center for Genetics (
http://research.marshfieldclinic.org/genetics/Map\_Markers/maps/IndexMapFrames.html
). The CMT1A/HNPP genomic region is shown as a shaded vertical bar. (▾) The predicted position of the centromere.
Figure 4
Genomic structure of the genes in the CMT1A/HNPP region. (A) PMP22. Arrowhead indicates marker D17S918, which contains polymorphic CAG repeats. Distances for each intron are shown at bottom, whereas individual exons are numbered at top. We hypothesized that in rare families with CMT accompanied by anticipation, it may be related to an expanded allele of this triplet repeat. To test this hypothesis, we obtained DNA samples from one such family for which the disease locus mapped to 17p11.2–17p12 by linkage analysis (Kovach et al. 1999). We examined the number of CAG repeat in members of this family, but failed to identify any expansion of the triplet repeats in affected individuals (data not shown). A point mutation in PMP22 was subsequently found to segregate with the disease phenotype in this family (Kovach et al. 1999). (B) HS3ST3B1 and HS3ST3A1. HS3STB1 is located inside the CMT1A/HNPP genomic region, whereas HS3STA1 is 569-kb telomeric to the distal CMT1A–REP. Each exon of the two genes is indicated as a box, and arrows show the direction of the transcription. (C) CDRT1 and NPD008/CGI-148. The horizontal shaded rectangle indicates the proximal CMT1A–REP. An open box shows the single exon of CDRT1 and solid boxes indicate seven exons of NPD008/CGI-148 that span 26 kb. (D) TEKT3 contains eight exons (solid boxes) including an untranslated first exon. The putative initiating methionine is in exon II. The exon/intron boundaries from exons III to VI, which were not determined by database analyses, were experimentally confirmed by RT–PCR and sequence analyses using single-stranded testis cDNA as the template DNA. (E) CDRT15 in the 11-kb low-copy repeat unit, LCRA1. Hatched horizontal bars indicate repetitive elements. Pseudogene for KIAA1151 is also shown.
Figure 5
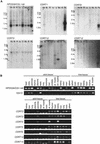
Expression studies of genes identified in the CMT1A/HNPP region. (A) Multiple-tissue Northern blot analyses. Tissues are indicated at top of each lane (He) Heart; (Br) brain; (Pl) placenta; (Lu) lung; (Li) liver; (Mu) skeletal muscle; (Ki) kidney; (Pa) pancreas. Marker sizes are 9.5, 7.5, 4.4, 2.4, and 1.35 kb. (B) Multiple-tissue RT–PCR analyses. NP008/CGI-148 is expressed in a wide variety of tissues. A 2-kb major transcript and two minor transcripts are observed. TEKT3 revealed no expression by Northern blotting (data not show), but high expression in testis by RT–PCR. Faint expression is also observed in both ovary and pancreas by RT–PCR. Fetal tissues reveal low level but ubiquitous expression. CDRT1 shows a major 2-kb and a minor 1-kb transcripts in pancreas. Faint expression is also identified in the heart. CDRT8 reveals two short transcript (0.7 and 1 kb) in the pancreas. A 3-kb transcript of CDRT9 represents low-level ubiquitous expression. CDRT10 is expressed in the skeletal muscle as a 1.3-kb transcript. CDRT12 reveals a 2.8-kb transcript in the pancreas at very low expression levels. CDRT2, CDRT3, CDRT4, CDRT5, CDRT6, CDRT14, and CDRT15 reveal no expression in adult tissues by Northern blotting (data not shown) and RT–PCR analyses, but show obvious expression in fetal tissues.
Figure 6
Gene evolution surrounding the proximal CMT1A–REP region. (A) A comparison of EST contigs between mouse and human. Eight mouse ESTs (shown as solid horizontal bars; GenBank accession nos. AI606691, AA089107, AA982328, AI181334, AI882388, AI429210, AI447154, and AI506955, from top to bottom) were reconstructed into a 1.5-kb contig with a 24-bp gap (horizontal rectangle with gradient colors) that aligns with two human genes, HREP (the numbers represent each exon of HREP; the exon VI does not align with the mouse EST contig) and CDRT1. Between the alignment of these two genes, there is a 269-bp region in the mouse clone that does not match any human sequence (purple arrow). The conceptual translation of this region does not identify a known protein functional motif. (B) A model for the evolution of new genes and the genomic structure surrounding the proximal CMT1A–REP region. Top figure represents the genomic structure of a hypothetical ancient gene AGIP (
A
ncestral
G
ene before the
I
ntegration of
P
roximal CMT1A–REP) modeled in mice. One or more exons originally contained in the AGIP are predicted to be lost by the integration of the proximal CMT1A–REP. Bottom figure shows human genomic structure in which HREP and CDRT1 are separated by the inserted proximal CMT1A–REP (dark rectangle). The pseudoexon of COX10 is utilized as the last exon of HREP from the opposite direction (green box).
Similar articles
- Charcot-Marie-Tooth disease and related inherited neuropathies.
Murakami T, Garcia CA, Reiter LT, Lupski JR. Murakami T, et al. Medicine (Baltimore). 1996 Sep;75(5):233-50. doi: 10.1097/00005792-199609000-00001. Medicine (Baltimore). 1996. PMID: 8862346 Review. - Novel PCR-based diagnostic tools for Charcot-Marie-Tooth type 1A and hereditary neuropathy with liability to pressure palsies.
Stronach EA, Clark C, Bell C, Löfgren A, McKay NG, Timmerman V, Van Broeckhoven C, Haites NE. Stronach EA, et al. J Peripher Nerv Syst. 1999;4(2):117-22. J Peripher Nerv Syst. 1999. PMID: 10442687 - Recombination hot spot in a 3.2-kb region of the Charcot-Marie-Tooth type 1A repeat sequences: new tools for molecular diagnosis of hereditary neuropathy with liability to pressure palsies and of Charcot-Marie-Tooth type 1A. French CMT Collaborative Research Group.
Lopes J, LeGuern E, Gouider R, Tardieu S, Abbas N, Birouk N, Gugenheim M, Bouche P, Agid Y, Brice A. Lopes J, et al. Am J Hum Genet. 1996 Jun;58(6):1223-30. Am J Hum Genet. 1996. PMID: 8651299 Free PMC article. - Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17.
Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR. Chance PF, et al. Hum Mol Genet. 1994 Feb;3(2):223-8. doi: 10.1093/hmg/3.2.223. Hum Mol Genet. 1994. PMID: 8004087 - Molecular mechanisms for CMT1A duplication and HNPP deletion.
Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR. Boerkoel CF, et al. Ann N Y Acad Sci. 1999 Sep 14;883:22-35. Ann N Y Acad Sci. 1999. PMID: 10586226 Review.
Cited by
- Anesthetic Considerations for Patients with Hereditary Neuropathy with Liability to Pressure Palsies: A Narrative Review.
Laudanski K, Elmadhoun O, Mathew A, Kahn-Pascual Y, Kerfeld MJ, Chen J, Sisniega DC, Gomez F. Laudanski K, et al. Healthcare (Basel). 2024 Apr 19;12(8):858. doi: 10.3390/healthcare12080858. Healthcare (Basel). 2024. PMID: 38667620 Free PMC article. Review. - AAV-mediated editing of PMP22 rescues Charcot-Marie-Tooth disease type 1A features in patient-derived iPS Schwann cells.
Yoshioka Y, Taniguchi JB, Homma H, Tamura T, Fujita K, Inotsume M, Tagawa K, Misawa K, Matsumoto N, Nakagawa M, Inoue H, Tanaka H, Okazawa H. Yoshioka Y, et al. Commun Med (Lond). 2023 Nov 28;3(1):170. doi: 10.1038/s43856-023-00400-y. Commun Med (Lond). 2023. PMID: 38017287 Free PMC article. - Bioinformatic analysis the expression and clinical significance of CDRT15 in cholangiocarcinoma using TCGA database.
Yu T, Zhang T, Zhao L, Li K, Li J, Yu A. Yu T, et al. Medicine (Baltimore). 2023 Aug 4;102(31):e34602. doi: 10.1097/MD.0000000000034602. Medicine (Baltimore). 2023. PMID: 37543771 Free PMC article. - Copy number variants from 4800 exomes contribute to ~7% of genetic diagnoses in movement disorders, muscle disorders and neuropathies.
Pennings M, Meijer RPP, Gerrits M, Janssen J, Pfundt R, de Leeuw N, Gilissen C, Gardeitchik T, Schouten M, Voermans N, van de Warrenburg B, Kamsteeg EJ. Pennings M, et al. Eur J Hum Genet. 2023 Jun;31(6):654-662. doi: 10.1038/s41431-023-01312-0. Epub 2023 Feb 13. Eur J Hum Genet. 2023. PMID: 36781956 Free PMC article. - Biology in balance: human diploid genome integrity, gene dosage, and genomic medicine.
Lupski JR. Lupski JR. Trends Genet. 2022 Jun;38(6):554-571. doi: 10.1016/j.tig.2022.03.001. Epub 2022 Apr 18. Trends Genet. 2022. PMID: 35450748 Free PMC article. Review.
References
- Badano, J.L., Inoue, K., Katsanis, N., and Lupski, J.R. New polymorphic short tandem repeats for PCR-based Charcot-Marie-Tooth disease type 1A duplication diagnosis. Clin. Chem. (In press). - PubMed
- Blair IP, Kennerson ML, Nicholson GA. Detection of Charcot-Marie-Tooth type 1A duplication by the polymerase chain reaction. Clin Chem. 1995;41:1105–1108. - PubMed
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR. Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann NY Acad Sci. 1999;883:22–35. - PubMed
- Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous