In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair - PubMed (original) (raw)
In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair
V Burdett et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Biochemical studies with model DNA heteroduplexes have implicated RecJ exonuclease, exonuclease VII, exonuclease I, and exonuclease X in Escherichia coli methyl-directed mismatch correction. However, strains deficient in the four exonucleases display only a modest increase in mutation rate, raising questions concerning involvement of these activities in mismatch repair in vivo. The quadruple mutant deficient in the four exonucleases, as well as the triple mutant deficient in RecJ exonuclease, exonuclease VII, and exonuclease I, grow poorly in the presence of the base analogue 2-aminopurine, and exposure to the base analogue results in filament formation, indicative of induction of SOS DNA damage response. The growth defect and filamentation phenotypes associated with 2-aminopurine exposure are effectively suppressed by null mutations in mutH, mutL, mutS, or uvrD/mutU, which encode activities that act upstream of the four exonucleases in the mechanism for the methyl-directed reaction that has been proposed based on in vitro studies. The quadruple exonuclease mutant is also cold-sensitive, having a severe growth defect at 30 degrees C. This phenotype is suppressed by a uvrD/mutU defect, and partially suppressed by mutH, mutL, or mutS mutations. These observations confirm involvement of the four exonucleases in methyl-directed mismatch repair in vivo and suggest that the low mutability of exonuclease-deficient strains is a consequence of under recovery of mutants due to a reduction in viability and/or chromosome loss associated with activation of the mismatch repair system in the absence of RecJ exonuclease, exonuclease VII, exonuclease I, and exonuclease X.
Figures
Figure 1
Suppression of 2APur sensitivity by inactivation of mismatch repair functions. LB agar plates containing 2APur (350 μg/ml) were spotted with 10-fold serial dilutions of each culture such that spots contained roughly 102–104 colony-forming units as indicated. Plates were photographed after 24 h at 37°C. Strain designations (Table 1) are shown on the left and relevant genotypes on the right. The small variability in cell densities evident is due to variation in the initial culture cell densities. In the absence of drug (data not shown), plating efficiency and colony size were similar for all strains tested.
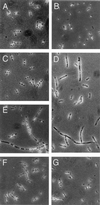
Figure 2
Cell and nucleoid morphologies associated with response to 2APur. Samples were removed from cultures grown overnight at 37°C in enriched minimal medium in the presence (350 μg/ml) or absence of 2APur and fixed on microscope slides. After staining with 4′,6-diamidino-2-phenylindole, cells were visualized and photographed by using phase-contrast and fluorescence optics as described in_Experimental Procedures_. All photographs are shown at the same magnification (×1,250). (A) BT199 no 2APur; (B) VB31 no 2APur; (C) BT199 plus 2APur; (D) VB31 plus 2APur; (E) STL4150 plus 2APur; (F) STL4150_mutS_ plus 2APur; (G) VB31_mutL_ plus 2APur.
Figure 3
Cold sensitivity of a RecJ− ExoVII− ExoI− ExoX− strain and suppression by mismatch repair defects. Two independent cultures of the strains shown (Table 1) were grown overnight in LB at 37°C, and 10-fold serial dilutions from each were prepared and spotted onto LB agar plates. One set of plates was photographed after incubation at 30°C for 24 h and the second after incubation for 42 h. Although not shown, growth at 37°C was comparable for all strains and was similar to that shown here for BT199. The approximate number of colony-forming units is indicated above each spot.
Similar articles
- Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair.
Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Viswanathan M, et al. J Biol Chem. 2001 Aug 17;276(33):31053-8. doi: 10.1074/jbc.M105481200. Epub 2001 Jun 19. J Biol Chem. 2001. PMID: 11418610 - Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance.
Viswanathan M, Lovett ST. Viswanathan M, et al. Genetics. 1998 May;149(1):7-16. doi: 10.1093/genetics/149.1.7. Genetics. 1998. PMID: 9584082 Free PMC article. - Methyl-directed DNA mismatch correction.
Modrich P. Modrich P. J Biol Chem. 1989 Apr 25;264(12):6597-600. J Biol Chem. 1989. PMID: 2651430 Review. - Mechanisms of DNA-mismatch correction.
Grilley M, Holmes J, Yashar B, Modrich P. Grilley M, et al. Mutat Res. 1990 Sep-Nov;236(2-3):253-67. doi: 10.1016/0921-8777(90)90009-t. Mutat Res. 1990. PMID: 2144613 Review. No abstract available.
Cited by
- Expanded roles for the MutL family of DNA mismatch repair proteins.
Furman CM, Elbashir R, Alani E. Furman CM, et al. Yeast. 2021 Jan;38(1):39-53. doi: 10.1002/yea.3512. Epub 2020 Jul 30. Yeast. 2021. PMID: 32652606 Free PMC article. Review. - RecJ-like protein from Pyrococcus furiosus has 3'-5' exonuclease activity on RNA: implications for proofreading of 3'-mismatched RNA primers in DNA replication.
Yuan H, Liu XP, Han Z, Allers T, Hou JL, Liu JH. Yuan H, et al. Nucleic Acids Res. 2013 Jun;41(11):5817-26. doi: 10.1093/nar/gkt275. Epub 2013 Apr 19. Nucleic Acids Res. 2013. PMID: 23605041 Free PMC article. - Epigenetic competition reveals density-dependent regulation and target site plasticity of phosphorothioate epigenetics in bacteria.
Wu X, Cao B, Aquino P, Chiu TP, Chen C, Jiang S, Deng Z, Chen S, Rohs R, Wang L, Galagan JE, Dedon PC. Wu X, et al. Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14322-14330. doi: 10.1073/pnas.2002933117. Epub 2020 Jun 9. Proc Natl Acad Sci U S A. 2020. PMID: 32518115 Free PMC article. - Structure and function of TatD exonuclease in DNA repair.
Chen YC, Li CL, Hsiao YY, Duh Y, Yuan HS. Chen YC, et al. Nucleic Acids Res. 2014;42(16):10776-85. doi: 10.1093/nar/gku732. Epub 2014 Aug 11. Nucleic Acids Res. 2014. PMID: 25114049 Free PMC article. - Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein.
Brown LT, Sutera VA Jr, Zhou S, Weitzel CS, Cheng Y, Lovett ST. Brown LT, et al. PLoS Genet. 2015 Nov 6;11(11):e1005651. doi: 10.1371/journal.pgen.1005651. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26544712 Free PMC article.
References
- Modrich P. Annu Rev Genet. 1991;25:229–253. - PubMed
- Radman M, Matic I, Halliday J A, Taddei F. Philos Trans R Soc London B. 1995;347:97–103. - PubMed
- Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. - PubMed
- Meselson M. In: Recombination of the Genetic Material. Low K B, editor. San Diego: Academic; 1988. pp. 91–113.
- Su S-S, Lahue R S, Au K G, Modrich P. J Biol Chem. 1988;263:6829–6835. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM023719/GM/NIGMS NIH HHS/United States
- R01 GM043889/GM/NIGMS NIH HHS/United States
- GM23719/GM/NIGMS NIH HHS/United States
- T32 GM07122/GM/NIGMS NIH HHS/United States
- GM43889/GM/NIGMS NIH HHS/United States
- T32 GM007122/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases