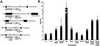Regulated transposition of a fish transposon in the mouse germ line - PubMed (original) (raw)
Regulated transposition of a fish transposon in the mouse germ line
S E Fischer et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Tc1/mariner elements are able to transpose in species other than the host from which they were isolated. As potential vectors for insertional mutagenesis and transgenesis of the mouse, these cut-and-paste transposons were tested for their ability to transpose in the mouse germ line. First, the levels of activity of several Tc1/mariner elements in mammalian cells were compared; the reconstructed fish transposon Sleeping Beauty (SB) was found to be an order of magnitude more efficient than the other tested transposons. SB then was introduced into the mouse germ line as a two-component system: one transgene for the expression of the transposase in the male germ line and a second transgene carrying a modified transposon. In 20% of the progeny of double transgenic male mice the transposon had jumped from the original chromosomal position into another locus. Analysis of the integration sites shows that these jumps indeed occurred through the action of SB transposase, and that SB has a strong preference for intrachromosomal transposition. Analysis of the excision sites suggests that double-strand breaks in haploid spermatids are repaired via nonhomologous end joining. The SB system may be a powerful tool for transposon mutagenesis of the mouse germ line.
Figures
Figure 1
Tc1/mariner transposons compared for their activity in HeLa cells. (A) Structure of the transposons tested and of the corresponding transposases (below the transposon). The black boxes represent the terminal inverted repeats. The diagonal lines indicate the positions of the SV40-G418-resistance cassette. The transposon sequence between these lines was deleted. The sizes of the wild-type and, parenthetically, disrupted transposons, are indicated. Each tagged transposon was cloned into the polycloning site in pUC18/19. The gray box in each the transposase protein represents the (predicted) DNA-binding domain, whereas the white part represents the catalytic domain containing the catalytic amino acids DD(34)E or DD(34)D(50). Relevant mutions are indicated beneath each transposon. All five ORFs have comparable numbers of optimal and nonoptimal human codons and are preceded and followed by identical sequences. (B) Activity of Tc1/mariner elements compared in HeLa cells. A vector with the transposon marked by an SV40-G418-resistance cassette was cotransfected with a CMV expression vector with either the ORF of the corresponding transposase or with the ORF of β-galactosidase. G418-resistant colonies were selected and counted. The transposition activity (indicated on the y axis) is the ratio of the number of resistant colonies obtained when the transposase expression vector is cotransfected over the number of resistant colonies when the β-galactosidase expression vector is cotransfected (1 indicates no transposition). The activity indicates an average of between three and nine independent transfections. The error bars indicate SEM.
Figure 2
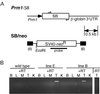
Generation and analysis of the transgenic mice. (A) Structure of the transgenes. The_Prm1-SB_-β -globin transgene is a fragment of plasmid pRP1345. A fragment of the mouse protamine 1 (Prm1) promoter was cloned in front of the ORF encoding the SB transposase, followed by the rabbit β-globin polyadenylation signal. The SB/neo transgene is a fragment of plasmid pT/neo. It has an SV40-G418R cassette in between the inverted repeats of the transposon. The fragments were injected into different FVB/N oocytes. Five founders (designated as lines A–E) transmitting the Prm1_-SB transgene to their offspring were obtained, ranging in copy number from 1 to 5. One line carrying a single copy of the complete SB/neo transposon was obtained (line 1).(B)_ RT-PCR analysis of the expression of SB transposase mRNA in several tissues, including brain (B), liver (L), skeletal muscle (M), testis (T) and kidney (K) of wild-type and transgenic mice (shown are lines B and E) using primers indicated in A by arrows.
Figure 3
(A) Mating scheme to induce and detect SB/neo transposition in mouse spermatids. Transposition events that occur in the spermatids of double transgenic males can be detected in their progeny. The progeny are analyzed for the presence of the SB/neo transposon by PCR. SB/neo+ animals (boxed) are further analyzed by PCR for the presence of SB/neo at the original locus and by Southern blotting to detect possible transposition events (shown in_B_). Males with SB/neo at a new location were further crossed to wild-type mice. SB/neo+ progeny (boxed) were analyzed to detect possible transposition events (shown in_D_). As an example, the lineage of animal 1734 is indicated (black text boxes). The heterozygous mice with an SB insertion were not homozygosed and had no obvious phenotypes. (B) Southern blot analysis of_Eco_RI-digested tail tip DNA of_Prm1-_SB+, SB/neo+ fathers and their offspring. The Eco_RI site in SB/neo and the probe, a fragment of the neo-resistance gene, are indicated in Fig. 2_A. Animal 1047 is the SB/neo+ founder animal (line 1) and shows the SB/neo transgene at its original locus. This line was crossed to all five_Prm1-_SB lines, lines A to E. Double transgenic males of lines 1A, 1B, 1C, 1D, and 1E were obtained and crossed with wild-type females. Of their progeny, animal 1724 did not inherit the SB/neo transgene; animals 1726 and 1728 did inherit the SB/neo transgene at its original locus. The remaining progeny represent 11 of 20 animals in which the SB/neo transposon had moved from its original locus to a new site (as the change in fragment size indicates). PCR analysis using a primer flanking the transposon at its original locus and a primer in the transposon confirmed the presence or absence of the transposon at the original locus. (C) Map of mouse chromosome 5 indicating the SB insertion sites. The insertion sites were mapped between the markers indicated, using the T31 mouse/hamster radiation hybrid panel. The distance between the original site on distal chromosome 5 and the region on central chromosome 5 is ≈25 cM. (D) Southern blot analysis of_Eco_RI-digested tail tip DNA of a_Prm1-_SB+, SB/neo+ father and his offspring. Animal 1047 is the SB/neo+ founder animal (line 1). Animal 1734 is progeny of a double transgenic male (line 1A), which has SB/neo at a new site and inherited the_Prm1-_SB transgene. Six progeny of mouse 1734 crossed to a wild-type female are shown: animals 1866, 1872, and 1874 have the transposon at the location of their father, animal 1734. Animal 1869 did not inherit the transposon. In animals 1867 and 1868 new transposition events were detected. PCR analysis confirmed the presence or absence of the transposon at the 1734 locus in the progeny of animal 1734. The positions of the fragments containing SB at its original site in animal 1047, and at the site in animal 1734, are indicated by arrows.
Similar articles
- Sleeping Beauty Transposition.
Ivics Z, Izsvák Z. Ivics Z, et al. Microbiol Spectr. 2015 Apr;3(2):MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. Microbiol Spectr. 2015. PMID: 26104705 Review. - Transposon mutagenesis of the mouse germline.
Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Carlson CM, et al. Genetics. 2003 Sep;165(1):243-56. doi: 10.1093/genetics/165.1.243. Genetics. 2003. PMID: 14504232 Free PMC article. - Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates.
Izsvák Z, Ivics Z, Plasterk RH. Izsvák Z, et al. J Mol Biol. 2000 Sep 8;302(1):93-102. doi: 10.1006/jmbi.2000.4047. J Mol Biol. 2000. PMID: 10964563 - Transposable elements for transgenesis and insertional mutagenesis in vertebrates: a contemporary review of experimental strategies.
Ivics Z, Izsvák Z. Ivics Z, et al. Methods Mol Biol. 2004;260:255-76. doi: 10.1385/1-59259-755-6:255. Methods Mol Biol. 2004. PMID: 15020812 - The Sleeping Beauty transposable element: evolution, regulation and genetic applications.
Ivics Z, Kaufman CD, Zayed H, Miskey C, Walisko O, Izsvák Z. Ivics Z, et al. Curr Issues Mol Biol. 2004 Jan;6(1):43-55. Curr Issues Mol Biol. 2004. PMID: 14632258 Review.
Cited by
- Advances in transposable elements: from mechanisms to applications in mammalian genomics.
Han M, Perkins MH, Novaes LS, Xu T, Chang H. Han M, et al. Front Genet. 2023 Nov 30;14:1290146. doi: 10.3389/fgene.2023.1290146. eCollection 2023. Front Genet. 2023. PMID: 38098473 Free PMC article. Review. - Jumping Ahead with Sleeping Beauty: Mechanistic Insights into Cut-and-Paste Transposition.
Ochmann MT, Ivics Z. Ochmann MT, et al. Viruses. 2021 Jan 8;13(1):76. doi: 10.3390/v13010076. Viruses. 2021. PMID: 33429848 Free PMC article. Review. - In vivo functional screening for systems-level integrative cancer genomics.
Weber J, Braun CJ, Saur D, Rad R. Weber J, et al. Nat Rev Cancer. 2020 Oct;20(10):573-593. doi: 10.1038/s41568-020-0275-9. Epub 2020 Jul 7. Nat Rev Cancer. 2020. PMID: 32636489 Review. - Targeting CD33 in Chemoresistant AML Patient-Derived Xenografts by CAR-CIK Cells Modified with an Improved SB Transposon System.
Rotiroti MC, Buracchi C, Arcangeli S, Galimberti S, Valsecchi MG, Perriello VM, Rasko T, Alberti G, Magnani CF, Cappuzzello C, Lundberg F, Pande A, Dastoli G, Introna M, Serafini M, Biagi E, Izsvák Z, Biondi A, Tettamanti S. Rotiroti MC, et al. Mol Ther. 2020 Sep 2;28(9):1974-1986. doi: 10.1016/j.ymthe.2020.05.021. Epub 2020 May 30. Mol Ther. 2020. PMID: 32526203 Free PMC article. - Jump around: transposons in and out of the laboratory.
Kumar A. Kumar A. F1000Res. 2020 Feb 24;9:F1000 Faculty Rev-135. doi: 10.12688/f1000research.21018.1. eCollection 2020. F1000Res. 2020. PMID: 32148769 Free PMC article. Review.
References
- Vos J C, de Baere I, Plasterk R H A. Genes Dev. 1996;10:755–761. - PubMed
- Catteruccia F, Nolan T, Loukeris T G, Blass C, Savakis C, Kafatos F C, Crisanti A. Nature (London) 2000;405:959–962. - PubMed
- Raz E, Van Luenen H G A M, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials