The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family - PubMed (original) (raw)
The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family
K L Hari et al. Genes Dev. 2001.
Abstract
The conserved heterochromatic location of centromeres in higher eukaryotes suggests that intrinsic properties of heterochromatin are important for chromosome inheritance. Based on this hypothesis, mutations in Drosophila melanogaster that alter heterochromatin-induced gene silencing were tested for effects on chromosome inheritance. Here we describe the characterization of the Su(var)2-10 locus, initially identified as a Suppressor of Position-Effect Variegation. Su(var)2-10 is required for viability, and mutations cause both minichromosome and endogenous chromosome inheritance defects. Mitotic chromosomes are improperly condensed in mutants, and polytene chromosomes are structurally abnormal and disorganized in the nucleus. Su(var)2-10 encodes a member of the PIAS protein family, a group of highly conserved proteins that control diverse functions. SU(VAR)2-10 proteins colocalize with nuclear lamin in interphase, and little to no SU(VAR)2-10 is found on condensed mitotic chromosomes. SU(VAR)2-10 is present at some polytene chromosome telomeres, and FISH analyses in mutant polytene nuclei revealed defects in telomere clustering and telomere-nuclear-lamina associations. We propose that Su(var2-10 controls multiple aspects of chromosome structure and function by establishing/maintaining chromosome organization in interphase nuclei.
Figures
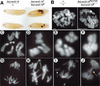
Figure 1
Chromosome structure and function defects in Su(var)2-10 homozygotes. (A) Melanotic tumors are visible in homozygous third-instar larvae (right side), but not heterozygous larvae (left side). (B) Mutant salivary gland nuclei (right panel) are significantly smaller and contain disorganized polytene chromosomes in comparison to wild-type nuclei (left panel). Note the absence of banding in the mutant nuclei. Bar, 5 μm. (C_–_J) DAPI-stained mitotic chromosome preparations from larval neuroblasts. Top panels are metaphase (C_–_F), bottom panels are in anaphase (G_–_J). C and G are controls, the rest are _Su(var)2-10 trans_-heterozygotes (see Materials and Methods). Note the abnormal chromosome condensation in D_—_F, and the aberrant segregation (H), chromosome fragments (I, arrow), and anaphase bridges (J, arrow) in mutant cells. The magnification for all mitotic figures is 1250×.
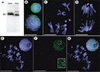
Figure 2
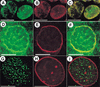
SU(VAR)2-10 localization in third-instar larval neuroblasts and S2 cells. (A) Western analysis of affinity-purified anti-SU(VAR)2-10. The antibodies recognize three distinct bands in a cytoplasmic extract (cyto) from 0–12-h embryos, and at least four bands in a nuclear extract (nuc) from the same developmental stage. (B_–_G) Indirect immunofluorescence showing SU(VAR)2-10 localization in mitotic cells. Green represents SU(VAR)2-10 protein, and blue DAPI-stained DNA. (B_–_D) In larval neuroblasts, SU(VAR)2-10 does not localize to chromosomes during metaphase (C) or anaphase (D), and is only prominent in interphase nuclei (B), where it is organized around the nuclear periphery and in numerous intranuclear spots. Bar, 5 μm. (E_–_G) Volume view of interphase and metaphase nuclei from S2 cells. (E) DNA alone, (F) SU(VAR)2-10 alone, and (G) the merged image. SU(VAR)2-10 antibodies stain interphase nuclei, but are not concentrated on metaphase chromosomes (F,G). Note the threadlike localization pattern in interphase cells (inset in F). Bar, 10 μm.
Figure 3
SU(VAR)2-10 does not overlap with GAGA factor in whole-mount larval salivary gland nuclei. Indirect immunofluorescence showing SU(VAR)2-10 and GAGA-factor localization in whole-mount salivary gland nuclei from third-instar larvae. In all panels, green is anti-SU(VAR)2-10 staining, red anti-GAGA staining, and blue DAPI-stained DNA. (A_–_C) Images from a single 0.2-μm optical section from the z-series stack of the deconvolved image. GAGA factor binds to discrete sites along polytene chromosomes (A), and does not overlap with anti-SU(VAR)2-10 staining (C). SU(VAR)2-10 is predominantly present in the interchromosomal spaces of the nucleoplasm and near the nuclear periphery, and weakly stains chromosomes (B). Bar, 15 μm. (D_–_F) SU(VAR)2-10 and GAGA-factor localization in whole-mount salivary gland nuclei from Su(var)2-101/Su(var)2-102 mutant larvae. (D) Nuclei are reduced in size and chromosome morphology is abnormal in mutant nuclei, yet GAGA factor is still able to associate with the DNA in a banded pattern. (E) SU(VAR)2-10 staining is decreased both in the cytoplasm and the nucleus. Bar, 10 μm.
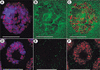
Figure 4
SU(VAR)2-10 is generally associated with squashed polytene chromosomes and does not colocalize with HP1 at telomeres. Indirect immunofluorescence showing SU(VAR)2-10 (green) on squashed polytene chromosomes (DAPI-stained DNA is blue in all panels). (A) SU(VAR)2-10 associates with polytene chromosomes in a general punctate pattern. The chromocenter is labeled (arrow), as are some euchromatic bands (arrowheads), and the tips of chromosomes 2L (B) and 4 (C), Bar in A, 15 μm. (B) Magnified view of area boxed in A showing that SU(VAR)2-10 (green) does not colocalize with HP1 (red) at the tip of chromosome 2L. (C) HP1 (red) densely stains the chromocenter and chromosome 4, and SU(VAR)2-10 (green) is found near the tip of chromosome 4. Bar, 10 μm. (D,E) SU(VAR)2-10 proteins localize distinctly to telomeres.
Figure 5
SU(VAR)2-10 and lamin colocalize around the nuclear periphery in interphase neuroblasts and salivary gland cells. Indirect immunofluorescence showing simultaneous localization of SU(VAR)2-10 and nuclear lamin in interphase cells from third-instar larval neuroblasts (A_–_C), whole-mount salivary gland nuclei (D_–_F), and interphase S2 cells (G_–_I). In all panels, green is anti-SU(VAR)2-10 staining and red is anti-lamin staining. SU(VAR)2-10 and lamin are tightly colocalized in neuroblasts (A_–_C) and salivary gland cells (D_–_F), but the two do not colocalize in S2 cells (G_–_I). Note the SU(VAR)2-10-specific staining in the interior of neuroblast nuclei (A,C). (A_–_C,G_–_I) Bar, 5 μm. (D_–_F) Bar, 15 μm.
Figure 6
SU(VAR)2-10 antibodies disrupt chromosome condensation and segregation in early embryos. Chromosome condensation and segregation are disrupted in embryos injected with anti-SU(VAR)2-10 antibodies (middle panels), but not in embryos injected with BSA (left panels) or α-amanitin (right panels), an RNA polymerase inhibitor. Embryos shown are at equivalent time points during the second mitotic cycle after injection. Time in minutes (t) postinjection is indicated for each panel. Note that at t = 25 metaphase chromosomes in the BSA- and α-amanitin-injected embryos are properly condensed and aligned at the metaphase plate. These nuclei divide and continue to cycle. During the second postinjection cycle in anti-SU(VAR)2-10-injected embryos, condensed metaphase chromosomes are never formed, and the mitotic cycles arrest prior to metaphase.
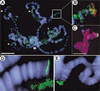
Figure 7
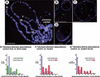
Defects in telomere–telomere and telomere–lamina associations occur in Su(var)2-10 mutant cells. FISH analyses using centric (green) and telomeric (red) probes to analyze chromosome organization in interphase polytene nuclei are shown. (A) Squashed polytene chromosomes from larval salivary glands were hybridized with the DNA probes and counterstained with DAPI (blue) to visualize DNA. Centric probes bind the heterochromatic chromocenter as expected, and telomeric probes bind the tips of chromosomes 2L and 3L Bar, 15 μm. Telomeric hybridization signals are separated in whole-mount nuclei from first-instar larval salivary glands (B), but are more tightly associated in nuclei from third-instar larvae (C). Telomere–telomere and telomere–lamina associations are disrupted in Su(var)2-10 mutant nuclei (D). Blue represents nuclear lamin staining in B_–_D Bar, 10 μm. (E_–_G) Bar graphs representing telomere–telomere and telomere–lamina distances relative to nuclear diameter for wild-type first-instar (green bars) and third-instar (blue bars) larvae, and for Su(var)2-10 mutant larvae (red bars). Two-sample _t_-tests confirmed that the differences between control and mutant ratios were statistically significant (see text for _p_-values).
Figure 8
Models for SU(VAR)2-10 function. In a wild-type interphase cell, SU(VAR)2-10 proteins (green balls) are present in the cytoplasm and the nucleus, where the predominant localization observed is near the nuclear periphery and in interchromosomal spaces (pink tubes, chromosomes; blue tubes, heterochromatin; wide blue tube, centromere). (A) SU(VAR)2-10 isoforms may function in a variety of transcriptional regulation complexes, associating with a number of chromosome-bound transcription factors (dark blue and purple ovals), and each association may control a different cellular response (e.g., viability, gene expression, or chromosome inheritance). (B) SU(VAR)2-10 coordinates chromosome organization in interphase nuclei, ensuring normal viability, gene expression, and chromosome inheritance. Importantly, the two models presented here are not mutually exclusive.
Similar articles
- Telomeric position effect--a third silencing mechanism in eukaryotes.
Doheny JG, Mottus R, Grigliatti TA. Doheny JG, et al. PLoS One. 2008;3(12):e3864. doi: 10.1371/journal.pone.0003864. Epub 2008 Dec 5. PLoS One. 2008. PMID: 19057646 Free PMC article. - SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation.
Cléard F, Delattre M, Spierer P. Cléard F, et al. EMBO J. 1997 Sep 1;16(17):5280-8. doi: 10.1093/emboj/16.17.5280. EMBO J. 1997. PMID: 9311988 Free PMC article. - Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation.
Spierer A, Seum C, Delattre M, Spierer P. Spierer A, et al. J Cell Sci. 2005 Nov 1;118(Pt 21):5047-57. doi: 10.1242/jcs.02623. Epub 2005 Oct 18. J Cell Sci. 2005. PMID: 16234327 - Histone modification and the control of heterochromatic gene silencing in Drosophila.
Ebert A, Lein S, Schotta G, Reuter G. Ebert A, et al. Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review. - SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.
Schotta G, Ebert A, Reuter G. Schotta G, et al. Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review.
Cited by
- Hippo pathway and Bonus control developmental cell fate decisions in the Drosophila eye.
Zhao H, Moberg KH, Veraksa A. Zhao H, et al. Dev Cell. 2023 Mar 13;58(5):416-434.e12. doi: 10.1016/j.devcel.2023.02.005. Epub 2023 Mar 2. Dev Cell. 2023. PMID: 36868234 Free PMC article. - SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes.
Zhao X. Zhao X. Mol Cell. 2018 Aug 2;71(3):409-418. doi: 10.1016/j.molcel.2018.07.027. Mol Cell. 2018. PMID: 30075142 Free PMC article. Review. - Identification of JAK/STAT pathway regulators--insights from RNAi screens.
Müller P, Boutros M, Zeidler MP. Müller P, et al. Semin Cell Dev Biol. 2008 Aug;19(4):360-9. doi: 10.1016/j.semcdb.2008.06.001. Epub 2008 Jun 8. Semin Cell Dev Biol. 2008. PMID: 18586112 Free PMC article. Review. - Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation.
Nie M, Xie Y, Loo JA, Courey AJ. Nie M, et al. PLoS One. 2009 Jun 16;4(6):e5905. doi: 10.1371/journal.pone.0005905. PLoS One. 2009. PMID: 19529778 Free PMC article. - HP1 is distributed within distinct chromatin domains at Drosophila telomeres.
Frydrychova RC, Mason JM, Archer TK. Frydrychova RC, et al. Genetics. 2008 Sep;180(1):121-31. doi: 10.1534/genetics.108.090647. Epub 2008 Aug 24. Genetics. 2008. PMID: 18723888 Free PMC article.
References
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. - PubMed
- Andrew DJ, Scott MP. Immunological methods for mapping protein distributions on polytene chromosomes. Methods Cell Biol. 1994;44:353–370. - PubMed
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. - PubMed
- Aravind L, Koonin EV. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous