A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters - PubMed (original) (raw)
A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters
M Kitagawa et al. Mol Cell Biol. 2001 Jul.
Abstract
Mastermind (Mam) has been implicated as an important positive regulator of the Notch signaling pathway by genetic studies using Drosophila melanogaster. Here we describe a biochemical mechanism of action of Mam within the Notch signaling pathway. Expression of a human sequence related to Drosophila Mam (hMam-1) in mammalian cells augments induction of Hairy Enhancer of split (HES) promoters by Notch signaling. hMam-1 stabilizes and participates in the DNA binding complex of the intracellular domain of human Notch1 and a CSL protein. Truncated versions of hMam-1 that can maintain an association with the complex behave in a dominant negative fashion and depress transactivation. Furthermore, Drosophila Mam forms a similar complex with the intracellular domain of Drosophila Notch and Drosophila CSL protein during activation of Enhancer of split, the Drosophila counterpart of HES. These results indicate that Mam is an essential component of the transcriptional apparatus of Notch signaling.
Figures
FIG. 1
Primary structure of Mam proteins. (a) Primary sequences of DMam (GenBank accession number X54251) and hMam-1 (D83785). Sequences were aligned using the ClustalW algorithm. Identical amino acids are in shaded boxes. Similar amino acids are in open boxes. The basic and acidic regions are in labeled boxes. The residues converted in the deletion mutants of hMam-1 are indicated by arrowheads. The overall identity of hMam-1 and DMam is 22%. Identity over the most highly conserved basic region is 38%. (b) Arrangement of basic and acidic regions of the DMam and hMam-1 proteins.
FIG. 1
Primary structure of Mam proteins. (a) Primary sequences of DMam (GenBank accession number X54251) and hMam-1 (D83785). Sequences were aligned using the ClustalW algorithm. Identical amino acids are in shaded boxes. Similar amino acids are in open boxes. The basic and acidic regions are in labeled boxes. The residues converted in the deletion mutants of hMam-1 are indicated by arrowheads. The overall identity of hMam-1 and DMam is 22%. Identity over the most highly conserved basic region is 38%. (b) Arrangement of basic and acidic regions of the DMam and hMam-1 proteins.
FIG. 1
Primary structure of Mam proteins. (a) Primary sequences of DMam (GenBank accession number X54251) and hMam-1 (D83785). Sequences were aligned using the ClustalW algorithm. Identical amino acids are in shaded boxes. Similar amino acids are in open boxes. The basic and acidic regions are in labeled boxes. The residues converted in the deletion mutants of hMam-1 are indicated by arrowheads. The overall identity of hMam-1 and DMam is 22%. Identity over the most highly conserved basic region is 38%. (b) Arrangement of basic and acidic regions of the DMam and hMam-1 proteins.
FIG. 2
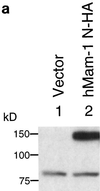
Identification and nuclear localization of the hMam-1 protein. (a) Identification of the hMam-1 protein. 293T cells were transfected with an expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS). Whole-cell extracts prepared from these cells were analyzed by immunoblotting using an anti-HA antibody. (b) Nuclear localization of the hMam-1 protein. 293T cells transfected with the expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS) were stained with the anti-HA antibody and a fluorescein isothiocyanate-conjugated secondary antibody. Phase-contrast, fluorescent, and merged views are shown.
FIG. 2
Identification and nuclear localization of the hMam-1 protein. (a) Identification of the hMam-1 protein. 293T cells were transfected with an expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS). Whole-cell extracts prepared from these cells were analyzed by immunoblotting using an anti-HA antibody. (b) Nuclear localization of the hMam-1 protein. 293T cells transfected with the expression vector for hMam-1 N-HA or the empty control vector (pEF-BOS) were stained with the anti-HA antibody and a fluorescein isothiocyanate-conjugated secondary antibody. Phase-contrast, fluorescent, and merged views are shown.
FIG. 3
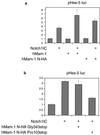
Effects of full-length and truncated forms of hMam-1 on transactivation of the HES-5 promoter. NIH 3T3 cells were transfected with expression vectors for Notch and Mam or their empty counterparts as controls. The transfection also contains the pHES-5 luciferase reporter and an internal control for transfection. Vertical axes represent normalized luciferase activity relative to the mean activity of empty vector-transfected cells. Error bars indicate standard deviations (n = 3). (a) hMam-1 and Notch1IC synergistically activate the target promoter. (b) hMam-1 truncated at proline residue 103 depresses the activity induced by Notch1IC.
FIG. 4
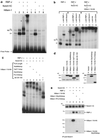
Formation of the Mam-Notch-CSL complex on its target promoter elements. (a) Extracts of transfected 293T cells exhibit activities that bind to the RBP-J element of the HES-1 promoter. Cells were transfected with the expression vectors for the indicated proteins. NS designates a nonspecific complex, open arrowheads mark RBP-J-specific bands, the closed arrowhead marks the RBP-J–Notch1IC-specific band, and stars mark hMam-1–RBP-J–Notch1IC-specific bands. The closed star marks the hMam-1–RBP-J–Notch1IC band that could be supershifted by the anti-Notch1 and anti-HA antibodies (see panel b). (b) Effects of antibodies on binding activities. Complexes formed in the presence of RBP-J, RBP-J–Notch1IC, or hMam-1–RBP-J–Notch1IC were tested for supershifts after incubation with the indicated antibodies. Anti-CD44 was included as a control. NS, arrowheads, and stars are as described for panel a. (c) hMam-1 and its truncations containing the basic charge cluster can associate with RBP-J and Notch1IC within ternary complexes. The hMam-1 truncations, characterized below, were tested for the ability to form the ternary complex that is observed with full-length hMam-1. 293T cells were transfected with the indicated combinations of the expression vectors, and their extracts were analyzed for binding to the RBP-J element. The binding complexes migrate at a rate proportional to the length of the truncated form of hMam-1. Stars are as described for panel a. (d) Expression of the truncated forms of the hMam-1 protein. Extracts of 293T cells transfected with the vectors for indicated proteins were blotted and stained with anti-HA antibody. (e) Expression of hMam-1 enhances the physical association of RBP-J with Notch1IC. 293T cell extracts transfected with indicated vectors were immunoprecipitated by anti-Notch1 antibody. The precipitates were separated on an SDS gel, blotted onto a membrane, and then stained sequentially with anti-Notch1, anti-RBP-J, and anti-HA antibodies. The hMam-1 N-HA Pro103stop protein was too small to be resolved on this gel.
FIG. 5
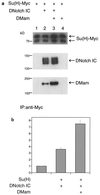
DMam function in cell culture. (a) Physical association of DMam with DNotchIC and Su(H). S2 cell extracts were immunoprecipitated with an anti-c-Myc antibody. The precipitates were resolved on an SDS gel, transferred to a membrane, and stained sequentially with anti-c-Myc, anti-Notch, and anti-DMam antibodies. Lane 2 shows that DMam is expressed endogenously in S2 cells. (b) DMam augments the activation of an E(spl) _m_γ reporter. The vertical axis represents normalized relative luciferase activity. The cotransfection of DMam with DNotch ICN (DNotchIC) and Su(H) leads to a doubling of reporter activity. Five experiments were performed, and all data was obtained in triplicate. Error bars indicate standard errors of the means.
Similar articles
- Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex.
Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Fryer CJ, et al. Genes Dev. 2002 Jun 1;16(11):1397-411. doi: 10.1101/gad.991602. Genes Dev. 2002. PMID: 12050117 Free PMC article. - Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors.
Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Wu L, et al. Mol Cell Biol. 2002 Nov;22(21):7688-700. doi: 10.1128/MCB.22.21.7688-7700.2002. Mol Cell Biol. 2002. PMID: 12370315 Free PMC article. - Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling.
Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. Lin SE, et al. J Biol Chem. 2002 Dec 27;277(52):50612-20. doi: 10.1074/jbc.M209529200. Epub 2002 Oct 16. J Biol Chem. 2002. PMID: 12386158 - Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex.
Kovall RA. Kovall RA. Curr Opin Struct Biol. 2007 Feb;17(1):117-27. doi: 10.1016/j.sbi.2006.11.004. Epub 2006 Dec 6. Curr Opin Struct Biol. 2007. PMID: 17157496 Review. - Notch signalling in the nucleus: roles of Mastermind-like (MAML) transcriptional coactivators.
Kitagawa M. Kitagawa M. J Biochem. 2016 Mar;159(3):287-94. doi: 10.1093/jb/mvv123. Epub 2015 Dec 28. J Biochem. 2016. PMID: 26711237 Review.
Cited by
- MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription.
Laduron S, Deplus R, Zhou S, Kholmanskikh O, Godelaine D, De Smet C, Hayward SD, Fuks F, Boon T, De Plaen E. Laduron S, et al. Nucleic Acids Res. 2004 Aug 17;32(14):4340-50. doi: 10.1093/nar/gkh735. Print 2004. Nucleic Acids Res. 2004. PMID: 15316101 Free PMC article. - High-resolution crystal structure of the human Notch 1 ankyrin domain.
Ehebauer MT, Chirgadze DY, Hayward P, Martinez Arias A, Blundell TL. Ehebauer MT, et al. Biochem J. 2005 Nov 15;392(Pt 1):13-20. doi: 10.1042/BJ20050515. Biochem J. 2005. PMID: 16011479 Free PMC article. - Transcription factor networks in Drosophila melanogaster.
Rhee DY, Cho DY, Zhai B, Slattery M, Ma L, Mintseris J, Wong CY, White KP, Celniker SE, Przytycka TM, Gygi SP, Obar RA, Artavanis-Tsakonas S. Rhee DY, et al. Cell Rep. 2014 Sep 25;8(6):2031-2043. doi: 10.1016/j.celrep.2014.08.038. Epub 2014 Sep 18. Cell Rep. 2014. PMID: 25242320 Free PMC article. - MAML1 enhances the transcriptional activity of Runx2 and plays a role in bone development.
Watanabe T, Oyama T, Asada M, Harada D, Ito Y, Inagawa M, Suzuki Y, Sugano S, Katsube K, Karsenty G, Komori T, Kitagawa M, Asahara H. Watanabe T, et al. PLoS Genet. 2013;9(1):e1003132. doi: 10.1371/journal.pgen.1003132. Epub 2013 Jan 10. PLoS Genet. 2013. PMID: 23326237 Free PMC article. - Delta-dependent Notch activation closes the early neuroblast temporal program to promote lineage progression and neurogenesis termination in Drosophila.
Sood C, Nahid MA, Branham KR, Pahl M, Doyle SE, Siegrist SE. Sood C, et al. Elife. 2024 Feb 23;12:RP88565. doi: 10.7554/eLife.88565. Elife. 2024. PMID: 38391176 Free PMC article.
References
- Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. - PubMed
- Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials