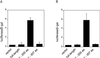Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation - PubMed (original) (raw)
Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation
O I Stenina et al. Proc Natl Acad Sci U S A. 2001.
Abstract
We have recently demonstrated that thrombin induces expression of the platelet-derived growth factor B-chain gene in endothelial cells (EC) through activation of the Y-box binding protein DNA-binding protein B (dbpB). We now present evidence that dbpB is activated by a novel mechanism: proteolytic cleavage leading to release from mRNA, nuclear translocation, and induction of thrombin-responsive genes. Cytosolic, full-length dbpB (50 kDa) was rapidly cleaved to a 30-kDa species upon thrombin stimulation of EC. This truncated, "active" dbpB exhibited nuclear localization and binding affinity for the thrombin response element sequence, which is distinct from the Y-box sequence. Oligo(dT) affinity chromatography revealed that cytosolic dbpB from control EC, but not active dbpB from thrombin-treated EC, was bound to mRNA. Latent dbpB immunoprecipitated from cytosolic extracts of control EC was activated by ribonuclease treatment. Furthermore, when EC cytosolic extracts were subjected to Nycodenz gradient centrifugation, latent dbpB fractionated with mRNA, whereas active dbpB fractionated with free proteins. The cytosolic retention domain of dbpB, which we localized to the region 247-267, was proteolytically cleaved during its activation. In contrast to full-length dbpB, truncated dbpB stimulated platelet-derived growth factor B-chain and tissue factor promoter activity by over 5-fold when transiently cotransfected with reporter constructs. These results suggest a novel mode of transcription factor activation in which an agonist causes release from mRNA of a latent transcription factor leading to its transport to the nucleus and its regulation of target gene expression.
Figures
Figure 1
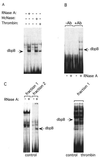
(A) Latent dbpB is associated with RNA in nonstimulated EC. Cytosolic extracts were prepared from either control bovine aortic EC or EC treated with bovine α-thrombin (10 units/ml, 2 h), as described in Experimental Procedures. Extracts from control EC were incubated with RNase A (1 mg/ml) or micrococcal nuclease (10,000 units/ml) for 30 min at 37°C before EMSA with the thrombin-response element (CCACCCACC) oligonucleotide as a probe. Arrow shows the position of dbpB activated by thrombin treatment of intact cells or ribonuclease treatment of extracts from untreated EC; McNase, micrococcal nuclease. (B) Latent dbpB coimmunoprecipitates with RNA. Cytosolic extracts prepared from control EC were incubated with preimmune serum or anti-dbpB antiserum raised against a C-terminal peptide of dbpB, followed by precipitation with protein A/G Sepharose. Protein was eluted by heat treatment. Aliquots of the eluants were treated with RNase A to release dbpB coimmunoprecipitated in association with RNA, and EMSA was performed. −Ab, preimmune serum used as a control; +Ab, immune serum used for precipitation of dbpB; RNase A, sample treated with ribonuclease A. (C) Oligo(dT) affinity chromatography of dbpB. (Left) EC cytosolic extracts were prepared from control and thrombin-treated EC as described in Experimental Procedures and dialyzed overnight in 0.2 M NaCl/0.01 M Tris⋅HCl, pH 7.5/0.002 M EDTA/0.2% Nonidet P-40/0.005 M β-mercaptoethanol. Oligo(dT)–biotin complex was added to the extract, and the mixture was applied to streptavidin magnetic beads. After rotation for 1 h at 4°C, the unbound fraction (fraction 1) was removed, the beads were washed three times, then washed with 25% formamide, and RNA was eluted with 60% formamide (fraction 2). The fractions were dialyzed overnight in EMSA buffer, and aliquots were treated with RNase A and subjected to EMSA. (Right) Fraction 1 from thrombin-treated EC lysates was used for EMSA without RNase treatment to detect activated dbpB.
Figure 2
Nycodenz gradient fractionation of cytosolic extracts from control and thrombin-stimulated EC. Lysates were prepared from nonstimulated or thrombin-stimulated EC and subjected to Nycodenz gradient fractionation as described in Experimental Procedures. Twenty-five fractions were collected from each tube, dialyzed, and used in EMSA after treatment with RNase to detect latent dbpB (A and D, fractions 15–21) or without any additional treatment (C) to detect thrombin-activated dbpB (C, fractions 6–10). Latent dbpB also was detected by Western blotting by using anti-dbpB antisera raised against the C-terminal peptide of dbpB (B). RNA isolation from fractions was performed by using Trizol reagent, and an oligo(dT) probe was used to detect mRNA by Northern blotting (not shown). Polysomal RNA (fractions 17–22) and 5S RNA (fractions 7–10) were detected by ethidium bromide staining of agarose gels (not shown). Free cytosolic protein (fractions 5–10) was detected by Coomassie staining of SDS/PAGE of individual fractions (not shown). C, cytosolic extract from unstimulated EC; T, cytosolic extract from thrombin-stimulated EC (arrow points to active dbpB in the extracts).
Figure 3
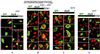
Identification of the cytosolic retention domain of dbpB. The fusion protein GFP-1–247 aa dbpB was localized to the nucleus (A). The addition of 20 aa (1–267 aa dbpB) resulted in its retention in the cytoplasm (B), similar to full-length dbpB (C). GFP-1–207 aa dbpB was localized to the nucleus (D) as well as the 191-, 200-, and 205-aa fusion proteins (not shown). Truncated dbpB mutants were prepared by introducing stop codons after the designated amino acid. Transient transfection of bovine aortic EC and the preparation of slides for fluorescence microscopy were performed as described (16).
Figure 4
Activation of PDGF and tissue-factor promoters by truncated dbpB. Truncated dbpB was prepared by introducing a stop codon into the cDNA after either 205 or 207 aa of the coding sequence. NIH 3T3 fibroblasts were transiently cotransfected with pcDNA3 containing the cDNAs for one of the dbpB species and either a 400-bp PDGF B-chain promoter–luciferase reporter construct (A) or a 660-bp tissue factor promoter–luciferase construct (B) as described in Experimental Procedures. Transfection (4 h) was followed by recovery in full growth medium for 36 h, at which time cells were lysed and assayed for luciferase activity. pβgal-control vector (CLONTECH) was cotransfected in all cases, and the activity of luciferase was normalized to β-galactosidase activity.
Figure 5
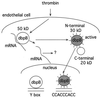
Model for agonist-induced dbpB activation. Latent dbpB bound to mRNA under basal conditions may act as a regulator of translation or as an RNA chaperone. Thrombin stimulation leads to dbpB cleavage, release from mRNA, translocation to the nucleus, and the regulation of thrombin-responsive gene expression (e.g., the PDGF B-chain gene). Truncated, activated dbpB has a distinct consensus binding site from that of full-length dbpB.
Similar articles
- Thrombin activates a Y box-binding protein (DNA-binding protein B) in endothelial cells.
Stenina OI, Poptic EJ, DiCorleto PE. Stenina OI, et al. J Clin Invest. 2000 Aug;106(4):579-87. doi: 10.1172/JCI9075. J Clin Invest. 2000. PMID: 10953033 Free PMC article. - Repression of major histocompatibility complex I-A beta gene expression by dbpA and dbpB (mYB-1) proteins.
Lloberas J, Maki RA, Celada A. Lloberas J, et al. Mol Cell Biol. 1995 Sep;15(9):5092-9. doi: 10.1128/MCB.15.9.5092. Mol Cell Biol. 1995. PMID: 7651426 Free PMC article. - Identification of a thrombin response element in the human platelet-derived growth factor B-chain (c-sis) promoter.
Scarpati EM, DiCorleto PE. Scarpati EM, et al. J Biol Chem. 1996 Feb 9;271(6):3025-32. doi: 10.1074/jbc.271.6.3025. J Biol Chem. 1996. PMID: 8621696 - Repression mechanisms of the I-A beta gene of the major histocompatibility complex.
Lloberas J, Soler C, Celada A. Lloberas J, et al. Immunobiology. 1997 Dec;198(1-3):249-63. doi: 10.1016/s0171-2985(97)80045-9. Immunobiology. 1997. PMID: 9442396 Review. - The role of nuclear Y-box binding protein 1 as a global marker in drug resistance.
Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi T, Iwamoto Y, Toi M, Fujii T, Yamana H, Kinoshita H, Kamura T, Tsuneyoshi M, Yasumoto K, Kohno K. Kuwano M, et al. Mol Cancer Ther. 2004 Nov;3(11):1485-92. Mol Cancer Ther. 2004. PMID: 15542787 Review.
Cited by
- Activated Protein C Ameliorates Renal Ischemia-Reperfusion Injury by Restricting Y-Box Binding Protein-1 Ubiquitination.
Dong W, Wang H, Shahzad K, Bock F, Al-Dabet MM, Ranjan S, Wolter J, Kohli S, Hoffmann J, Dhople VM, Zhu C, Lindquist JA, Esmon CT, Gröne E, Gröne HJ, Madhusudhan T, Mertens PR, Schlüter D, Isermann B. Dong W, et al. J Am Soc Nephrol. 2015 Nov;26(11):2789-99. doi: 10.1681/ASN.2014080846. Epub 2015 May 26. J Am Soc Nephrol. 2015. PMID: 26015455 Free PMC article. - β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis.
Sulatskaya AI, Kosolapova AO, Bobylev AG, Belousov MV, Antonets KS, Sulatsky MI, Kuznetsova IM, Turoverov KK, Stepanenko OV, Nizhnikov AA. Sulatskaya AI, et al. Int J Mol Sci. 2021 Oct 20;22(21):11316. doi: 10.3390/ijms222111316. Int J Mol Sci. 2021. PMID: 34768745 Free PMC article. Review. - PDGFR-α inhibition preserves blood-brain barrier after intracerebral hemorrhage.
Ma Q, Huang B, Khatibi N, Rolland W 2nd, Suzuki H, Zhang JH, Tang J. Ma Q, et al. Ann Neurol. 2011 Dec;70(6):920-31. doi: 10.1002/ana.22549. Ann Neurol. 2011. PMID: 22190365 Free PMC article. - YB-1 is a Transcription/Translation Factor that Orchestrates the Oncogenome by Hardwiring Signal Transduction to Gene Expression.
Wu J, Stratford AL, Astanehe A, Dunn SE. Wu J, et al. Transl Oncogenomics. 2007 May 11;2:49-65. Print 2007. Transl Oncogenomics. 2007. PMID: 23641145 Free PMC article. - Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1.
Safak M, Sadowska B, Barrucco R, Khalili K. Safak M, et al. J Virol. 2002 Apr;76(8):3828-38. doi: 10.1128/jvi.76.8.3828-3838.2002. J Virol. 2002. PMID: 11907223 Free PMC article.
References
- Darnell J E., Jr J Interferon Cytokine Res. 1998;18:549–554. - PubMed
- Boulikas T. Crit Rev Eukaryotic Gene Expression. 1995;5:1–77. - PubMed
- Meek D W. Cell Signalling. 1998;10:159–166. - PubMed
- Ghosh S, Baltimore D. Nature (London) 1990;344:678–682. - PubMed
- Wang X, Sato R, Brown M S, Hua X, J L, Goldstein J L. Cell. 1994;77:53–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources