Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p - PubMed (original) (raw)
Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p
J Walter et al. EMBO J. 2001.
Abstract
Tail-anchored proteins are distinct from other membrane proteins as they are thought to insert into the endoplasmic reticulum (ER) membrane independently of Sec61p translocation pores. These pores not only mediate import but are also assumed to catalyze export of proteins in a process called ER-associated protein degradation (ERAD). In order to examine the Sec61p dependence of the export of tail-anchored proteins, we analyzed the degradation pathway of a tail-anchored ER membrane protein, the ubiquitin-conjugating enzyme 6 (Ubc6p). In contrast to other ubiquitin conjugating enzymes (Ubcs), Ubc6p is naturally short-lived. Its proteolysis is mediated specifically by the unique Ubc6p tail region. Degradation further requires the activity of Cue1p-assembled Ubc7p, and its own catalytic site cysteine. However, it occurs independently of the other ERAD components Ubc1p, Hrd1p/Der3p, Hrd3p and Der1p. In contrast to other natural ERAD substrates, proteasomal mutants accumulate a membrane-bound degradation intermediate of Ubc6p. Most interestingly, mutations in SEC61 do not reduce the turnover of full-length Ubc6p nor cause a detectable accumulation of degradation intermediates. These data are in accordance with a model in which tail-anchored proteins can be extracted from membranes independently of Sec61p.
Figures
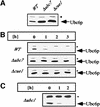
Fig. 1. Ubc6p degradation depends on Ubc7p and Cue1p, but noton Ubc1p. (A) Immunoblots loaded with equal amounts of isolated membranes from the strains indicated were probed with αUbc6p antibody. (B) The stability of Ubc6p was analyzed in cycloheximide decay experiments of the same strains: after abrogation of protein translation by the addition of cycloheximide, aliquots were taken at the indicated time points, and membranes were prepared and probed for the amount of Ubc6p. (C) Cycloheximide decay experiment in a strain depleted of UBC1. The asterisk marks Sec61p, which served as a loading control.
Fig. 2. Ubc6p degradation is mediated by its tail domain. The stability of Ubc4p, Ubc6p–TM and Ubc4ptail was tested in cycloheximide decay experiments and analyzed by immunoblotting. Experiments were conducted as in Figure 1B, except that in the upper two panels total cell lysates were probed. Ubc4p was tested in a wild-type strain. Ubc6p deleted for its transmembrane anchor (Ubc6p–TM) and the Ubc4p–Ubc6ptail fusion (Ubc4ptail) were expressed from a low-copy plasmid in a Δubc6 strain. The asterisk marks Sec61p, which served as a loading control.
Fig. 3. Ubc6p degradation depends on polyubiquitylation, catalytically active Ubc7p and its own activity. Cycloheximide decay experiments conducted as in Figure 1B and analyzed by immunoblotting. (A and B) The genomic UBC7 and UBC6 genes were mutated in their active site cysteine resulting in _ubc7_ser and _ubc6_ser, which were tested for the stability of Ubc6p and Ubc6pser, respectively. (C) The HA-tagged Ubc6p (Ubc6pha) was expressed from a low-copy plasmid in a wild-type strain and in the _ubc6_ser strain. The asterisk marks Sec61p, which served as a loading control. (D) The dependence of Ubc6p on polyubiquitylation was tested by the co-expression of mutant ubiquitinK48R. (E) Δubc6 cells were transformed with high-copy plasmids coding for Ubc6p, Ubc6pser or an empty vector. Microsome preparations from equal amounts of cells were immunoprecipitated with anti-Ubc6p antibody, and western blots were analyzed with an anti-ubiquitin antibody. The Ubc6p immunoprecipitation was controlled by blotting aliquots for Ubc6p (data not shown).
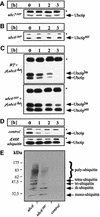
Fig. 4. Proteasomal mutants stabilize Ubc6p and cause the accumulation of a degradation intermediate. (A) Cycloheximide decay experiment of the proteasomal mutant strain pre1-1, conducted as in Figure 1B and analyzed by immunoblotting. A fragment accumulated, which reacted with the αUbc6p antibody. (B) Western blot of membrane preparations from the strains indicated. (C) Western blot of total cell lysates from the strains indicated. (D) Membranes from the pre1-1 strain were extracted with agents that strip peripheral proteins from the membranes, and with detergent, as indicated. Equal amounts of supernatant (sn) and pellet (p) were blotted for Ubc6p. The asterisk marks Sec61p, which served as a loading control.
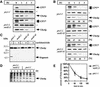
Fig. 5. Ubc6p degradation does not depend on Hrd1p/Der3p, Hrd3p or Der1p. (A) The immunoblots show cycloheximide decay experiments, conducted as in Figure 1B, of Δhrd1/der3, Δhrd3 and Δder1 deletion strains and of the corresponding wild-type strains. All strains expressed the prc1-1 allele, which codes for the classical ERAD substrate CPY*. It served as a control for the stabilizing effects on ERAD. The asterisk (*) marks Sec61p, which served as a loading control. (B) Quantification of the luminescence of the Ubc6p signals in an experiment as in (A).
Fig. 6. Sec61p function in Ubc6p degradation. (A and B) Cycloheximide decay experiment conducted in sec61 mutant strains and the corresponding wild type. Membranes were immunoblotted and probed for the indicated proteins. The asterisk marks Sec61p, which served as a loading control. The experiments in (A) and (B) were conducted at 25°C. CPY* demonstrates the translocation defects of these strains. The lower band reacting with the αCPY antibody indicates the non-glycosylated precursor, which results from an impaired import. The amount of Ubc6p was determined using a luminescence imager. The relative intensity is given as a percentage of the value at the zero time point. (C) The indicated yeast strains treated with cycloheximide for 3 h at 25°C. Aliquots before and after cycloheximide treatment were taken and membranes immunoblotted for Ubc6p degradation intermediates. (D) Radioactive pulse–chase experiment of the same strains as in (A) after a 1 h pre-shift to 37°C. Total cell lysates were immuno precipitated for Ubc6p. (E) Quantification of experiments as shown in (D). Three independent experiments were averaged and the standard deviation was calculated and is indicated by error bars (values for prc1-1 are 100%; 45.1 ± 1.9%; 17.5 ± 3.6%; 12.0 ± 1.7%; and for prc1-1, sec61-2 are 100%; 43.4 ± 10.8%; 21.3 ± 6.1%; 14.5 ± 1.7%).
Similar articles
- Role of Cue1p in ubiquitination and degradation at the ER surface.
Biederer T, Volkwein C, Sommer T. Biederer T, et al. Science. 1997 Dec 5;278(5344):1806-9. doi: 10.1126/science.278.5344.1806. Science. 1997. PMID: 9388185 - Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation.
Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Bays NW, et al. Nat Cell Biol. 2001 Jan;3(1):24-9. doi: 10.1038/35050524. Nat Cell Biol. 2001. PMID: 11146622 - Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair.
Gilon T, Chomsky O, Kulka RG. Gilon T, et al. Mol Cell Biol. 2000 Oct;20(19):7214-9. doi: 10.1128/MCB.20.19.7214-7219.2000. Mol Cell Biol. 2000. PMID: 10982838 Free PMC article. - Endoplasmic reticulum degradation: reverse protein flow of no return.
Sommer T, Wolf DH. Sommer T, et al. FASEB J. 1997 Dec;11(14):1227-33. doi: 10.1096/fasebj.11.14.9409541. FASEB J. 1997. PMID: 9409541 Review. - The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications.
Ciechanover A, Orian A, Schwartz AL. Ciechanover A, et al. J Cell Biochem Suppl. 2000;34:40-51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. J Cell Biochem Suppl. 2000. PMID: 10762014 Review.
Cited by
- Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope.
Zattas D, Hochstrasser M. Zattas D, et al. Crit Rev Biochem Mol Biol. 2015 Jan-Feb;50(1):1-17. doi: 10.3109/10409238.2014.959889. Epub 2014 Sep 18. Crit Rev Biochem Mol Biol. 2015. PMID: 25231236 Free PMC article. Review. - Selective clearance of aberrant membrane proteins by TORC1-mediated micro-ER-phagy.
Gyurkovska V, Alvarado Cartagena YM, Murtazina R, Zhao SF, Ximenez de Olaso C, Segev N. Gyurkovska V, et al. Cell Rep. 2025 Feb 25;44(2):115282. doi: 10.1016/j.celrep.2025.115282. Epub 2025 Feb 12. Cell Rep. 2025. PMID: 39946230 Free PMC article. - Doa1 targets ubiquitinated substrates for mitochondria-associated degradation.
Wu X, Li L, Jiang H. Wu X, et al. J Cell Biol. 2016 Apr 11;213(1):49-63. doi: 10.1083/jcb.201510098. Epub 2016 Apr 4. J Cell Biol. 2016. PMID: 27044889 Free PMC article. - Degradation of integral membrane proteins modified with the photosensitive degron module requires the cytosolic endoplasmic reticulum-associated degradation pathway.
Scheffer J, Hasenjäger S, Taxis C. Scheffer J, et al. Mol Biol Cell. 2019 Sep 15;30(20):2558-2570. doi: 10.1091/mbc.E18-12-0754. Epub 2019 Aug 14. Mol Biol Cell. 2019. PMID: 31411939 Free PMC article. - Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation.
Kostova Z, Tsai YC, Weissman AM. Kostova Z, et al. Semin Cell Dev Biol. 2007 Dec;18(6):770-9. doi: 10.1016/j.semcdb.2007.09.002. Epub 2007 Sep 8. Semin Cell Dev Biol. 2007. PMID: 17950636 Free PMC article. Review.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Stuhl,K. (1989) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY.
- Bays N.W., Gardener,R.G., Seelig,L.P., Joazeiro,C.A. and Hampton,R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. - PubMed
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous