The 19S complex of the proteasome regulates nucleotide excision repair in yeast - PubMed (original) (raw)
The 19S complex of the proteasome regulates nucleotide excision repair in yeast
T G Gillette et al. Genes Dev. 2001.
Abstract
Previous studies suggest that the amino-terminal ubiquitin-like (ubl) domain of Rad23 protein can recruit the proteasome for a stimulatory role during nucleotide excision repair in the yeast Saccharomyces cerevisiae. In this report, we show that the 19S regulatory complex of the yeast proteasome can affect nucleotide excision repair independently of Rad23 protein. Strains with mutations in 19S regulatory subunits (but not 20S subunits) of the proteasome promote partial recovery of nucleotide excision repair in vivo in rad23 deletion mutants, but not in other nucleotide excision repair-defective strains tested. In addition, a strain that expresses a temperature-degradable ATPase subunit of the 19S regulatory complex manifests a dramatically increased rate of nucleotide excision repair in vivo. These data indicate that the 19S regulatory complex of the 26S proteasome can negatively regulate the rate of nucleotide excision repair in yeast and suggest that Rad23 protein not only recruits the 19S regulatory complex, but also can mediate functional interactions between the 19S regulatory complex and the nucleotide excision repair machinery. The 19S regulatory complex of the yeast proteasome functions in nucleotide excision repair independent of proteolysis.
Figures
Figure 1
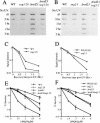
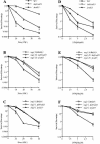
UV radiation sensitivity in sug1-2 and rad23 mutant strains. (▪) Wild-type strain Sc507 (A,B); Sc342 (C). (□) sug mutant strains sug1-20 (A); sug1-25 (B); sug2-1 (C). (▴) rad23 null strain. (▵) sug rad23 strain, in the same order as sug single mutant strains.
Figure 2
4NQO sensitivity and removal of photoproducts in vivo in sug1-2 and rad23 mutant strains. (A,B) Time course of (6-4) photoproduct lesion removal as detected by antibody. No UV indicates DNA from cells immediately before UV treatment and t0 immediately post UV treatment. All other points are time points of recovery at 30°C post-treatment. (WT) Parental strain for sug1-20 (A) and sug2-1 (B) strains. (C,D) Quantitation of slot blot. Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1. (E,F) 4NQO survival: (▪) wild-type strain Sc507 (E) and Sc342 (F); (□) sug mutant strains sug1-20 (E) and sug2-1 (F); (▴) rad23 null strain; (▵) sug rad23 strain, in the same order as sug single mutant strains.
Figure 3
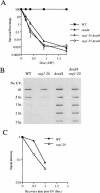
UV radiation sensitivity and NER in the sug1 rad4 double mutant. (A) UV survival curves: (▪) Sc507 wild-type strain; (▴) rad4 null mutant; (▵) sug1-20 rad4 double mutant; (○) sug1-25 rad4 strain. (B) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. (C) Quantitation of slot blot (in A). Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1.
Figure 4
UV radiation sensitivity and NER in the sug1 rad7 double mutant. (A): (▪) Sc507 wild-type strain; (▴) rad7 null mutant; (▵) sug1-20 rad7 double mutant; (○) sug1-25 rad7 strain. (B) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody.
Figure 5
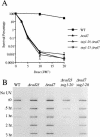
UV radiation sensitivity and NER in 20S proteasome mutants. (A) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. Method the same as in Fig. 2, except cells moved to 37°C for 90 min before UV treatment and subsequent recovery at 37°C post-UV treatment. (B) Quantitation of slot blot (in A). Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1. (C): (▪) Wild-type strain; (□) pre1-1 pre4-1 mutant; (⋄) pre1-1 pre2-2 mutant; (▴) rad23 null mutant; (▵) rad23 pre1-1 pre4-1 mutant strain; (○) rad23 pre1-1 pre2-2 strain.
Figure 6
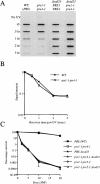
Sensitivity to UV radiation and 4NQO of strains carrying rad23 lacking the ubl domain. UV/4NQO survival curves. (A,D): (▪) Sc507 wild-type strain; (▴) rad23 null mutant; (●) rad23 null mutant carrying a centromeric plasmid expressing Δubl rad23. (B,E): (□) sug1-20 strain; (▵) sug1-20 rad23 mutant; (○) sug1-20 rad23 mutant carrying a centromeric plasmid expressing Δubl rad23. (C,F): (□) sug2-1 strain; (▵) sug2-1 rad23 mutant; (○) sug2-1 rad23 mutant carrying a centromeric plasmid expressing Δubl rad23.
Figure 7
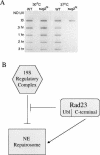
(A) Removal of photoproducts in vivo after degradation of Sug2 protein. Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. Strains where grown to mid-log phase at 30°C and then switched to the indicated temperature for 5 h before UV treatment as well as the post-treatment recovery period. (WT) Isogenic parent of the sug2ts strain. (B) Model of Rad23 and 19S genetic interactions in NER. In this model, the 19S regulatory complex of the proteasome is shown to negatively modulate the rate of NER in vivo. This effect is attenuated by interaction of the amino-terminal ubl domain of Rad23 protein. The carboxyl terminus of Rad23 enhances NER through direct interaction with other subunits of the nucleotide excision repairosome.
Similar articles
- The link between 20S proteasome activity and post-replication DNA repair in Saccharomyces cerevisiae.
Podlaska A, McIntyre J, Skoneczna A, Sledziewska-Gojska E. Podlaska A, et al. Mol Microbiol. 2003 Sep;49(5):1321-32. doi: 10.1046/j.1365-2958.2003.03635.x. Mol Microbiol. 2003. PMID: 12940990 - Proteasome subunit Rpn1 binds ubiquitin-like protein domains.
Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GA, Finley D. Elsasser S, et al. Nat Cell Biol. 2002 Sep;4(9):725-30. doi: 10.1038/ncb845. Nat Cell Biol. 2002. PMID: 12198498 - Rad23 links DNA repair to the ubiquitin/proteasome pathway.
Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Schauber C, et al. Nature. 1998 Feb 12;391(6668):715-8. doi: 10.1038/35661. Nature. 1998. PMID: 9490418 - The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation.
Dantuma NP, Heinen C, Hoogstraten D. Dantuma NP, et al. DNA Repair (Amst). 2009 Apr 5;8(4):449-60. doi: 10.1016/j.dnarep.2009.01.005. Epub 2009 Feb 14. DNA Repair (Amst). 2009. PMID: 19223247 Review. - A role for Rad23 proteins in 26S proteasome-dependent protein degradation?
van Laar T, van der Eb AJ, Terleth C. van Laar T, et al. Mutat Res. 2002 Jan 29;499(1):53-61. doi: 10.1016/s0027-5107(01)00291-3. Mutat Res. 2002. PMID: 11804604 Review.
Cited by
- UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair.
Sugasawa K. Sugasawa K. J Mol Histol. 2006 Sep;37(5-7):189-202. doi: 10.1007/s10735-006-9044-7. Epub 2006 Jul 21. J Mol Histol. 2006. PMID: 16858626 Review. - A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein.
Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. Ng JM, et al. Genes Dev. 2003 Jul 1;17(13):1630-45. doi: 10.1101/gad.260003. Epub 2003 Jun 18. Genes Dev. 2003. PMID: 12815074 Free PMC article. - Regulation of ubiquitin ligase dynamics by the nucleolus.
Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Mekhail K, et al. J Cell Biol. 2005 Aug 29;170(5):733-44. doi: 10.1083/jcb.200506030. J Cell Biol. 2005. PMID: 16129783 Free PMC article. - Catalytically Active Proteasomes Function Predominantly in the Cytosol.
Dang FW, Chen L, Madura K. Dang FW, et al. J Biol Chem. 2016 Sep 2;291(36):18765-77. doi: 10.1074/jbc.M115.712406. Epub 2016 Jul 14. J Biol Chem. 2016. PMID: 27417138 Free PMC article. - Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13.
Lundgren J, Masson P, Realini CA, Young P. Lundgren J, et al. Mol Cell Biol. 2003 Aug;23(15):5320-30. doi: 10.1128/MCB.23.15.5320-5330.2003. Mol Cell Biol. 2003. PMID: 12861018 Free PMC article.
References
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. - PubMed
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. - PubMed
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases