Molecular mechanisms of translation initiation in eukaryotes - PubMed (original) (raw)
Review
Molecular mechanisms of translation initiation in eukaryotes
T V Pestova et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Translation initiation is a complex process in which initiator tRNA, 40S, and 60S ribosomal subunits are assembled by eukaryotic initiation factors (eIFs) into an 80S ribosome at the initiation codon of mRNA. The cap-binding complex eIF4F and the factors eIF4A and eIF4B are required for binding of 43S complexes (comprising a 40S subunit, eIF2/GTP/Met-tRNAi and eIF3) to the 5' end of capped mRNA but are not sufficient to promote ribosomal scanning to the initiation codon. eIF1A enhances the ability of eIF1 to dissociate aberrantly assembled complexes from mRNA, and these factors synergistically mediate 48S complex assembly at the initiation codon. Joining of 48S complexes to 60S subunits to form 80S ribosomes requires eIF5B, which has an essential ribosome-dependent GTPase activity and hydrolysis of eIF2-bound GTP induced by eIF5. Initiation on a few mRNAs is cap-independent and occurs instead by internal ribosomal entry. Encephalomyocarditis virus (EMCV) and hepatitis C virus epitomize distinct mechanisms of internal ribosomal entry site (IRES)-mediated initiation. The eIF4A and eIF4G subunits of eIF4F bind immediately upstream of the EMCV initiation codon and promote binding of 43S complexes. EMCV initiation does not involve scanning and does not require eIF1, eIF1A, and the eIF4E subunit of eIF4F. Initiation on some EMCV-like IRESs requires additional noncanonical initiation factors, which alter IRES conformation and promote binding of eIF4A/4G. Initiation on the hepatitis C virus IRES is even simpler: 43S complexes containing only eIF2 and eIF3 bind directly to the initiation codon as a result of specific interaction of the IRES and the 40S subunit.
Figures
Figure 1
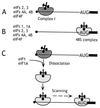
The mechanism of action of eIF1 and eIF1A in promoting assembly of 48S ribosomal complexes at the authentic initiation codon of a conventional capped mRNA. The 5′ terminal m7G residue is shown as a filled black circle, the 5′ NTR as a black line, and the ORF downstream of the AUG initiation codon as a black rectangle. (A) In the presence of eIFs 2, 3, 4A, 4B, and 4F, an aberrant ribosomal complex (“complex I”) assembles at a cap-proximal position but is unable to scan downstream to the initiation codon. (B) In the presence of eIFs 1, 1A, 2, 3, 4A, 4B, and 4F, 48S ribosomal complexes assemble exclusively at the authentic initiation codon. (C) Addition of eIF1 and eIF1A to complex I promotes its complete conversion to correctly assembled 48S complexes after dissociation of complex I and rebinding of 43S ribosomal complexes in a scanning-competent form.
Figure 2
Sequence and structural conservation of eukaryotic eIF5B proteins from Homo sapiens (15), Drosophila melanogaster (16), and Saccharomyces cerevisiae (14), archaeal IF2 from Methanococcus jannaschii and prokaryotic IF2 from Escherichia coli (12). The percentages of amino acid identities to human eIF5B in the N-terminal region of the protein, the GTP-binding domain, and the C-terminal region of the protein are shown. The black rectangle in the schematic representation identifies the position of the GTP-binding domains in these proteins with the indicated GTP-binding protein consensus sequence motifs G1-G5 aligned with sequence motifs G1-G5 of E. coli IF2 and human eIF5B. Numbers above the domains of eIF5B/IF2 proteins refer to the amino acid residues in each protein; numbers below the aligned sequences refer to the amino acid residues in G1–G5 motifs of human eIF5B.
Figure 3
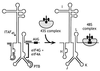
Schematic representation of EMCV, FMDV, and TMEV IRES domains H–L, showing binding sites for PTB (thick gray line) and ITAF45 (black icosahedron), as determined by footprinting (30, 31), and for the eIF4G/eIF4A complex, as determined by footprinting (31, 32) and toeprinting (26, 27, 33). The interaction of eIF4G/eIF4A with the J–K domain, which is essential for recruitment of the 43S complex to the initiation codon, is enhanced by PTB and ITAF45 in an IRES-specific manner, as discussed in the text.
Figure 4
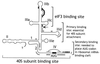
Schematic secondary structure of domains II, III, and IV of HCV-like IRESs, showing sites of interaction with eIF3 (thick black lines) (47) and with 40S subunits (thick gray lines) (48, 49). The toeprints detected at the leading edge of bound eIF3 (46, 47, 49, 50) are indicated by an arrow. The toeprints at the leading edge of 40S subunits in binary IRES:40S subunit complexes are indicated by open circles and in 48S complexes formed on inclusion of eIF2/GTP/Met-tRNAi with 40S subunits by filled circles (46, 50). Toeprints caused by ribosomal contact with the pseudoknot are not shown. Sequences flanking the initiation codon are base paired to form domain IV in HCV but not in BVDV and CSFV. BVDV and CSFV contain two hairpins (IIId1 and IIId2) at an analogous position to HCV IIId. The nomenclature of helices in the pseudoknot and of domains is as in ref. .
Similar articles
- Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP.
Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Unbehaun A, et al. Genes Dev. 2004 Dec 15;18(24):3078-93. doi: 10.1101/gad.1255704. Genes Dev. 2004. PMID: 15601822 Free PMC article. - Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site.
de Breyne S, Yu Y, Pestova TV, Hellen CU. de Breyne S, et al. RNA. 2008 Feb;14(2):367-80. doi: 10.1261/rna.696508. Epub 2007 Dec 19. RNA. 2008. PMID: 18094123 Free PMC article. - The joining of ribosomal subunits in eukaryotes requires eIF5B.
Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. Pestova TV, et al. Nature. 2000 Jan 20;403(6767):332-5. doi: 10.1038/35002118. Nature. 2000. PMID: 10659855 - The scanning mechanism of eukaryotic translation initiation.
Hinnebusch AG. Hinnebusch AG. Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review. - Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites.
Pisarev AV, Shirokikh NE, Hellen CU. Pisarev AV, et al. C R Biol. 2005 Jul;328(7):589-605. doi: 10.1016/j.crvi.2005.02.004. C R Biol. 2005. PMID: 15992743 Review.
Cited by
- The impact of the phosphomimetic eIF2αS/D on global translation, reinitiation and the integrated stress response is attenuated in N2a cells.
Legrand N, Jaquier-Gubler P, Curran J. Legrand N, et al. Nucleic Acids Res. 2015 Sep 30;43(17):8392-404. doi: 10.1093/nar/gkv827. Epub 2015 Aug 11. Nucleic Acids Res. 2015. PMID: 26264663 Free PMC article. - Translational Regulation by eIFs and RNA Modifications in Cancer.
Zhang L, Zhang Y, Zhang S, Qiu L, Zhang Y, Zhou Y, Han J, Xie J. Zhang L, et al. Genes (Basel). 2022 Nov 6;13(11):2050. doi: 10.3390/genes13112050. Genes (Basel). 2022. PMID: 36360287 Free PMC article. Review. - Evidence for a Negative Cooperativity between eIF5A and eEF2 on Binding to the Ribosome.
Rossi D, Barbosa NM, Galvão FC, Boldrin PE, Hershey JW, Zanelli CF, Fraser CS, Valentini SR. Rossi D, et al. PLoS One. 2016 Apr 26;11(4):e0154205. doi: 10.1371/journal.pone.0154205. eCollection 2016. PLoS One. 2016. PMID: 27115996 Free PMC article. - Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes.
Pfingsten JS, Costantino DA, Kieft JS. Pfingsten JS, et al. J Mol Biol. 2007 Jul 27;370(5):856-69. doi: 10.1016/j.jmb.2007.04.076. Epub 2007 May 8. J Mol Biol. 2007. PMID: 17544444 Free PMC article. - The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS. Monzingo AF, et al. Plant Physiol. 2007 Apr;143(4):1504-18. doi: 10.1104/pp.106.093146. Epub 2007 Feb 23. Plant Physiol. 2007. PMID: 17322339 Free PMC article.
References
- Pestova T V, Borukhov S I, Hellen C U T. Nature (London) 1998;394:854–859. - PubMed
- Chaudhuri J, Chowdhury D, Maitra U. J Biol Chem. 1999;274:17975–17980. - PubMed
- Battiste J L, Pestova T V, Hellen C U T, Wagner G. Mol Cell. 2000;5:109–119. - PubMed
- Dahlquist K D, Puglisi J D. J Mol Biol. 2000;299:1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous