Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips - PubMed (original) (raw)
. 2001 Jul;39(7):2531-40.
doi: 10.1128/JCM.39.7.2531-2540.2001.
S Lapa, D Gryadunov, A Sobolev, B Strizhkov, N Chernyh, O Skotnikova, O Irtuganova, A Moroz, V Litvinov, M Vladimirskii, M Perelman, L Chernousova, V Erokhin, A Zasedatelev, A Mirzabekov
Affiliations
- PMID: 11427565
- PMCID: PMC88181
- DOI: 10.1128/JCM.39.7.2531-2540.2001
Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips
V Mikhailovich et al. J Clin Microbiol. 2001 Jul.
Abstract
Three new molecular approaches were developed to identify drug-resistant strains of Mycobacterium tuberculosis using biochips with oligonucleotides immobilized in polyacrylamide gel pads. These approaches are significantly faster than traditional bacteriological methods. All three approaches-hybridization, PCR, and ligase detection reaction--were designed to analyze an 81-bp fragment of the gene rpoB encoding the beta-subunit of RNA polymerase, where most known mutations of rifampin resistance are located. The call set for hybridization analysis consisted of 42 immobilized oligonucleotides and enabled us to identify 30 mutant variants of the rpoB gene within 24 h. These variants are found in 95% of all mutants whose rifampin resistance is caused by mutations in the 81-bp fragment. Using the second approach, allele-specific on-chip PCR, it was possible to directly identify mutations in clinical samples within 1.5 h. The third approach, on-chip ligase detection reaction, was sensitive enough to reveal rifampin-resistant strains in a model mixture containing 1% of resistant and 99% of susceptible bacteria. This level of sensitivity is comparable to that from the determination of M. tuberculosis drug resistance by using standard bacteriological tests.
Figures
FIG. 1
The images (A and B) and intensities (C and D) of hybridization. The immobilized probes are listed in Table 1. (A and C) Wild-type target DNA; (B and D) His-526-Tyr mutant target DNA. The fluorescence intensities within each column were normalized to a maximal fluorescence signal corresponding to a perfect hybridization duplex. a. u., arbitrary units. The arrow in panel B points to the only gel pad where a perfect duplex formed, resulting in a higher level of fluorescence.
FIG. 2
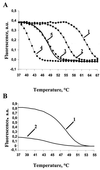
Melting curves of hybridization duplexes. (A) Effect of buffer composition on the melting temperature of a perfect duplex formed between oligonucleotide probe a3 (Table 1) and a PCR-amplified fragment of the wild-type rpoB gene (labeled target DNA). The samples were melted in 1 M NaCl (curve 1), 1 M NaCl with 10% formamide (curve 2), 20% formamide (curve 3), 30% formamide (curve 4), or 1 M GuCNS (curve 5). (B) Melting curves of perfect (curve 1) and imperfect (curve 2) duplexes in 1 M GuCNS. The duplexes were formed by a PCR-amplified fragment of the wild-type rpoB gene and oligonucleotide probe a3 (perfect duplex) or b3 (imperfect duplex). a. u., arbitrary units.
FIG. 3
Allele-specific on-chip PCR. A strong fluorescence signal is observed when the 3′ nucleotide of the immobilized primer is complementary to target DNA, extended by Taq polymerase, and forms a stable duplex with the fluorescently labeled target DNA. Immobilized primers are listed in Table 5. (A) Wild-type target DNA; (B) His-526-Asp mutant target DNA (CAC→GAC); the corresponding gel pad is marked with an arrow.
FIG. 4
Detection of mutant DNA by on-chip LDR. Each reaction mixture contained a total of 3 pmol of single-stranded DNA. Reaction a was performed with wild-type DNA; reaction e was performed with His-526-Leu mutant DNA; other reactions contained 3 pmol of wild-type DNA with a mixture of 1% (b), 2% (c), or 10% (d) of the mutant DNA.
References
- Bocharov P P. Series criterion based on median. In: Pechinkin A B, editor. Mathematical statistics. 1994. p. 164. RUDN, Moscow, Russia.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous