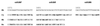Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus - PubMed (original) (raw)
Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus
Z Ge et al. J Clin Microbiol. 2001 Jul.
Abstract
Helicobacter hepaticus infection in mice is being used as an animal model for elucidating the pathogenesis of gastrointestinal and biliary diseases in humans. H. hepaticus, which forms a spreading film on selective agar, is not amenable to routine quantitative counts of organisms in tissues using a CFU method. In this study, a fluorogenic PCR-based assay was developed to quantitatively detect H. hepaticus in mouse ceca and feces using the ABI Prism 7700 sequence detection system. A pair of primers and a probe for this assay were generated from the H. hepaticus cdtB gene (encoding subunit B of the H. hepaticus cytolethal distending toxin). Using this assay, the sensitivity for detection of H. hepaticus chromosomal DNA prepared from pure culture was 20 fg, which is equivalent to approximately 14 copies of the H. hepaticus genome based on an estimated genome size of approximately 1.3 Mb determined by pulsed-field gel electrophoresis. H. hepaticus present in feces and cecal samples from H. hepaticus-infected mice was readily quantified. The selected PCR primers and probe did not generate fluorescent signals from eight other helicobacters (H. canis, H. cineadi, H. felis, H. mustelae, H. nemestrinae, H. pullorum, H. pylori, and H. rodentium). A fluorescent signal was detected from 20 ng of H. bilis DNA but with much lower sensitivity (10(6)-fold) than from H. hepaticus DNA. Therefore, this assay can be used with high sensitivity and specificity to quantify H. hepaticus in experimentally infected mouse models as well as in naturally infected mice.
Figures
FIG. 1
Sequence comparison between the H. hepaticus primers and probe and the corresponding regions of the cdtB genes in selected bacteria. Hh, H. hepaticus; Cj, C. jejuni; Hb, H. bilis; Hc, H. canis. Numbers indicate the positions of the respective primers and probe in the nucleotide sequence determined by Young et al. (25). The nucleotides different from those present in the H. hepaticus cdtB gene are indicated in boldface. The sequence of primer cdtBR is reverse and complementary to the sequence shown here. For H. bilis and H. canis, only the partial sequence of the cdtB gene is available (2).
FIG. 2
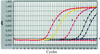
Detection limitation of the TaqMan PCR assay for H. hepaticus genomic DNA. Amplification plots 1 to 6 were generated from 2 × 107, 2 × 106, 2 × 104, 200, 20, and 2 fg, respectively. The _r_2 value from the linear regression in this assay is >0.99.
FIG. 3
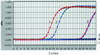
Evaluation of the specificity of the primers and probe for H. hepaticus. Amplification plots 1 and 2 were generated using 2 × 107 and 2 × 106 fg of H. hepaticus chromosomal DNA, respectively, whereas plots 3 to 11 were produced using 2 × 107 fg of chromosomal DNA from H. bilis, H. canis, H. cineadi, H. felis, H. mustelae, H. nemestrinae, H. pullorum, H. pylori, and H. rodentium, respectively.
FIG. 4
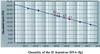
Quantitative detection of H. hepaticus present in mouse ceca and feces. The quantities of H. hepaticus genomic DNA used for the standard curve are indicated on the x axis, and the corresponding Ct values are given on the y axis. Twenty nanograms of total DNA from each sample was used in this assay. C1 (cecum) and F1 (feces), DNA from mouse I; C2 (cecum) and F2 (feces), DNA from mouse II.
References
- Chien C C, Taylor N S, Ge Z, Schauer D B, Young V B, Fox J G. Identification of cdtB homologues and cytolethal distending toxin activity in enterohepatic Helicobacter spp. J Med Microbiol. 2000;49:525–534. - PubMed
- Chin E Y, Dangler C A, Fox J G, Schauer D B. Helicobacter hepaticus infection triggers IBD in TCR alpha beta mutant mice. Comp Med. 2000;50:586–592. - PubMed
- Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous