Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair - PubMed (original) (raw)
Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair
M Missura et al. EMBO J. 2001.
Abstract
The multiprotein factor composed of XPA and replication protein A (RPA) is an essential subunit of the mammalian nucleotide excision repair system. Although XPA-RPA has been implicated in damage recognition, its activity in the DNA repair pathway remains controversial. By replacing DNA adducts with mispaired bases or non-hybridizing analogues, we found that the weak preference of XPA and RPA for damaged substrates is entirely mediated by indirect readout of DNA helix conformations. Further screening with artificially distorted substrates revealed that XPA binds most efficiently to rigidly bent duplexes but not to single-stranded DNA. Conversely, RPA recognizes single-stranded sites but not backbone bending. Thus, the association of XPA with RPA generates a double-check sensor that detects, simultaneously, backbone and base pair distortion of DNA. The affinity of XPA for sharply bent duplexes, characteristic of architectural proteins, is not compatible with a direct function during recognition of nucleotide lesions. Instead, XPA in conjunction with RPA may constitute a regulatory factor that monitors DNA bending and unwinding to verify the damage-specific localization of repair complexes or control their correct three-dimensional assembly.
Figures
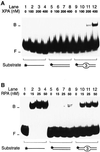
Fig. 1. Recognition of artificial DNA distortions. (A) Electrophoretic mobility shift assay demonstrating preferential binding of XPA protein to DNA duplexes containing, in the centre, three consecutive mis matches (lanes 9–12), but no binding to DNA single strands (lanes 1–4). The position of free (F) and bound (B) DNA is indicated. The asterisks denote a 32P label on the 5′ end of 19mer substrates. (B) Comparison with RPA: under identical reaction conditions, RPA retains its characteristic preference for single-stranded DNA (lanes 1–4).
Fig. 2. Synergic recognition of DNA distortions by XPA and RPA. (A) Electrophoretic mobility shift assay demonstrating synergic binding of XPA and RPA (100 nM each) to 43mer DNA duplexes containing three mismatches in the centre (lanes 9 and 10, in duplicate). The position of free DNA (F) and the position of nucleoprotein complexes with XPA and RPA are indicated. (B) Longer autoradiographic exposure of lanes 6–9, illustrating the different electrophoretic mobility of various complexes. The position of nucleoprotein complexes containing XPA, RPA or both XPA and RPA is indicated. (C) Quantitative evaluation of mobility shift assays (mean values of two experiments). Left panel: percentages of bound DNA obtained in the presence of the 43mer homoduplex control. Right panel: percentages of bound DNA upon incubation with 43mer DNA fragments containing three mismatches in the centre. Protein concentrations ranged from 100 to 300 nM.
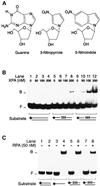
Fig. 3. Recognition of non-hybridizing base analogues. (A) Nucleoside analogues containing 3-nitropyrrole or 5-nitroindole. Non-hybridizing base analogues lack donor and acceptor groups for Watson–Crick hydrogen bonding. (B) Electrophoretic mobility shift assay demonstrating binding of XPA protein to DNA distortions generated by three consecutive 3-nitropyrroles (lanes 5–8) or three consecutive 5-nitroindoles (lanes 9–12). The asterisks denote a 32P label on the 5′ end of 19mer DNA. (C) Comparison with RPA under identical binding conditions. RPA was incubated with homoduplex DNA (lane 2), single-stranded DNA (lane 4) or duplexes containing, in the centre, either three 3-nitropyrroles (lane 6) or three 5-nitroindoles (lane 8).
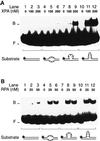
Fig. 4. Recognition of bulged DNA. (A) Electrophoretic mobility shift assay demonstrating the increased affinity of XPA for a single-stranded DNA loop (lanes 7–9) and, particularly, for a double-stranded loop in 19 bp DNA (lanes 10–12). The loop of lanes 7–9 results from the insertion of three unpaired nucleotides, while the loop of lanes 10–12 consists of three GC base pairs. The asterisks denote a 32P label on the 5′ end of each substrate. (B) Comparison with RPA: under identical reaction conditions, RPA binds to extra-helical loops (lanes 7–12) only slightly more efficiently than to three mismatches (lanes 4–6).
Fig. 5. XPA is an architectural protein that recognizes kinked backbones. (A) Electrophoretic mobility shift assay demonstrating the extraordinary affinity of XPA protein for synthetic three- (lanes 4–6) and four-way DNA junctions (lanes 7–9). (B) Comparison between Y-shaped DNA molecules (lanes 5–8) and three-way DNA junctions (lanes 9–12). (C) XPA can be cross-linked to four-way DNA junctions. Reaction mixtures containing DNA and the indicated concentrations of XPA were pre-incubated for 10 min in the dark, followed by a 20 min exposure to UV light (366 nm). Cross-linked samples were analysed on a denaturing polyacrylamide gel. The filled circle indicates the photoreactive 4-thio-deoxythimidine. (D) Mobility shift assay showing that RPA, unlike XPA, has no increased affinity for three-way junctions (lanes 5–8) or four-way junctions (lanes 9–12) compared with linear duplex DNA (lanes 1–4).
Fig. 6. Recognition and excision of GpG platinum cross-links. (A) Electrophoretic mobility shift assay in which XPA protein was incubated with 20mer duplexes containing a single GpG cross-link. The mononuclear cisplatin cross-link (Pt), which induces a rigid kink, is recognized (lanes 5–8), while the dinuclear analogue (Pt-Pt), in which this kink is replaced by a flexible hinge, is not recognized (lanes 9–12). (B) Comparison with RPA. Under identical reaction conditions, the mononuclear cisplatin cross-link (lanes 5–8) is recognized by RPA less efficiently than the dinuclear analogue (lanes 9–12). (C) Quantitative evaluation of two independent mobility shift assays performed with either XPA or RPA and platinated substrates. (D) Excision assay in HeLa cell extract demonstrating that both the mononuclear (lane 2) and the dinuclear GpG cross-link (lane 3) are repaired. The main excision products have a size of 29–30 nucleotides. Lane 1 shows a control reaction with undamaged DNA. The substrate preparations of lanes 1 and 3 were contaminated with a small fraction of short fragments (<148 nucleotides) resulting from incomplete oligonucleotide ligation.
Fig. 7. XPA–RPA generates dynamic complexes with NER substrates. (A) Nucleoprotein complexes were produced by incubating XPA protein (400 nM; lanes 1–4) or RPA (50 nM; lanes 5–8) with 19mer substrates containing a benzo[_a_]pyrene-dG adduct. Lanes 1 and 5 show the complexes formed during the first 10 min of incubation. After this pre-incubation time, the reactions were supplemented with a 100-fold excess of cold competitor DNA (19mer duplexes with three 5-nitroindole analogues in the centre). The samples were further incubated for the time intervals indicated and loaded on to the gel for electrophoresis. The stepwise appearance of the bands reflects the delay with which the samples were applied on to the running gel. (B) Nucleoprotein complexes produced by incubating XPA and RPA (100 nM each) with 43mer substrates containing three consecutive 5-nitroindoles in the centre. The band shifts formed by either factor alone are shown in lanes 1 (XPA) and 2 (RPA). Lane 3 demonstrates that, together, XPA and RPA bind all the DNA substrate during a pre-incubation of 10 min. The samples of lanes 4–6 were loaded after the time periods indicated, following the addition of a 100-fold excess of competitor DNA. (C) Excision assay in HeLa cell extract. The human NER system excises three consecutive 5-nitroindoles (lane 2). A single 5-nitroindole (lane 1) or undamaged DNA (lane 3) is not repaired. Lane 4 shows a control reaction with substrate containing a benzo[_a_]pyrene-dG adduct, confirming that the excision products are within the expected size range of 24–32 nucleotides.
Fig. 8. Inhibition of DNA unwinding. (A) ATP-independent unwinding of partial duplex DNA by RPA. The positions of circular substrate and of displaced radiolabelled primers are indicated. (B) RPA (200 nM) and partial duplex substrate were incubated with increasing concentrations of XPA protein. The sample of lane 9 was denatured by boiling for 5 min before electrophoretic analysis. (C) Quantitative evaluation of two independent unwinding assays indicating >50% inhibition at an XPA:RPA ratio of 1:1.
Similar articles
- Thermodynamic properties of damaged DNA and its recognition by xeroderma pigmentosum group A protein and replication protein A.
Brabec V, Stehlíková K, Malina J, Vojtiísková M, Kaspárková J. Brabec V, et al. Arch Biochem Biophys. 2006 Feb 1;446(1):1-10. doi: 10.1016/j.abb.2005.12.003. Epub 2005 Dec 22. Arch Biochem Biophys. 2006. PMID: 16405861 - RPA involvement in the damage-recognition and incision steps of nucleotide excision repair.
He Z, Henricksen LA, Wold MS, Ingles CJ. He Z, et al. Nature. 1995 Apr 6;374(6522):566-9. doi: 10.1038/374566a0. Nature. 1995. PMID: 7700386 - Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA.
Buschta-Hedayat N, Buterin T, Hess MT, Missura M, Naegeli H. Buschta-Hedayat N, et al. Proc Natl Acad Sci U S A. 1999 May 25;96(11):6090-5. doi: 10.1073/pnas.96.11.6090. Proc Natl Acad Sci U S A. 1999. PMID: 10339546 Free PMC article. - Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA.
Morikawa K, Shirakawa M. Morikawa K, et al. Mutat Res. 2000 Aug 30;460(3-4):257-75. doi: 10.1016/s0921-8777(00)00031-8. Mutat Res. 2000. PMID: 10946233 Review. - Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair.
Thoma BS, Vasquez KM. Thoma BS, et al. Mol Carcinog. 2003 Sep;38(1):1-13. doi: 10.1002/mc.10143. Mol Carcinog. 2003. PMID: 12949838 Review.
Cited by
- Metal nanoparticles for cancer therapy: Precision targeting of DNA damage.
Chen Q, Fang C, Xia F, Wang Q, Li F, Ling D. Chen Q, et al. Acta Pharm Sin B. 2024 Mar;14(3):1132-1149. doi: 10.1016/j.apsb.2023.08.031. Epub 2023 Sep 3. Acta Pharm Sin B. 2024. PMID: 38486992 Free PMC article. Review. - Melatonin improves the vitrification of sheep morulae by modulating transcriptome.
Ji P, Liu Y, Yan L, Jia Y, Zhao M, Lv D, Yao Y, Ma W, Yin D, Liu F, Gao S, Wusiman A, Yang K, Zhang L, Liu G. Ji P, et al. Front Vet Sci. 2023 Oct 18;10:1212047. doi: 10.3389/fvets.2023.1212047. eCollection 2023. Front Vet Sci. 2023. PMID: 37920328 Free PMC article. - A disease-associated XPA allele interferes with TFIIH binding and primarily affects transcription-coupled nucleotide excision repair.
van den Heuvel D, Kim M, Wondergem AP, van der Meer PJ, Witkamp M, Lambregtse F, Kim HS, Kan F, Apelt K, Kragten A, González-Prieto R, Vertegaal ACO, Yeo JE, Kim BG, van Doorn R, Schärer OD, Luijsterburg MS. van den Heuvel D, et al. Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2208860120. doi: 10.1073/pnas.2208860120. Epub 2023 Mar 9. Proc Natl Acad Sci U S A. 2023. PMID: 36893274 Free PMC article. - The XPA Protein-Life under Precise Control.
Krasikova YS, Lavrik OI, Rechkunova NI. Krasikova YS, et al. Cells. 2022 Nov 22;11(23):3723. doi: 10.3390/cells11233723. Cells. 2022. PMID: 36496984 Free PMC article. Review. - A nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance.
Li F, Sun H, Ren J, Zhang B, Hu X, Fang C, Lee J, Gu H, Ling D. Li F, et al. Nat Commun. 2022 Nov 30;13(1):7361. doi: 10.1038/s41467-022-35022-w. Nat Commun. 2022. PMID: 36450764 Free PMC article.
References
- Aboussekhra A. et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified components. Cell, 80, 859–868. - PubMed
- Araújo S.J., Tirode,F., Coin,F., Pospiech,H., Syväoja,J.E., Stucki,M., Hübscher,U., Egly,J.-M. and Wood,R.D. (2000) Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev., 14, 349–359. - PMC - PubMed
- Asahina H., Kuraoka,I., Shirakawa,M., Morita,E.H., Miura,N., Miyamoto,I., Ohtsuka,E., Okada,Y. and Tanaka,K. (1994) The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat. Res., 315, 229–237. - PubMed
- Batty D., Rapic’-Otrin,V., Levine,A.S. and Wood,R.D. (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol., 300, 275–290. - PubMed
- Burns J.L., Guzder,S.N., Sung,P., Prakash,S. and Prakash,L. (1996) An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem., 271, 11607–11610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources